Limitações termodinâmicas no surgimento natural de moléculas de cadeia longa: Implicações para a origem da vida
quinta-feira, julho 31, 2025
Darwin, nós temos um problema muito sério - já estão intitulando artigos com a palavra proibida: Design!
sexta-feira, junho 20, 2025
Cell
Volume 188, Issue 12p3202-3218.e21June 12, 2025
Design principles of cell-state-specific enhancers in hematopoiesis
Robert Frömel1,2 ∙ Julia Rühle1,2 ∙ Aina Bernal Martinez1 ∙ … ∙ Felix Pacheco Pastor1 ∙ Rosa Martinez-Corral3 ∙ Lars Velten1,2,4 et al
Highlights
• Cell-state-specific activity measurements of 64,400 minimalistic enhancers
• Individual transcription factors are often both activators and repressors
• Negative synergies between activators result in cell-state-specific repression
• Modeling enables the automated design of cell-type-specific enhancers in blood
Summary
During cellular differentiation, enhancers transform overlapping gradients of transcription factors (TFs) to highly specific gene expression patterns. However, the vast complexity of regulatory DNA impedes the identification of the underlying cis-regulatory rules. Here, we characterized 64,400 fully synthetic DNA sequences to bottom-up dissect design principles of cell-state-specific enhancers in the context of the differentiation of blood stem cells to seven myeloid lineages. Focusing on binding sites for 38 TFs and their pairwise interactions, we found that identical sites displayed both repressive and activating function as a consequence of cell state, site combinatorics, or simply predicted occupancy of a TF on an enhancer. Surprisingly, combinations of activating sites frequently neutralized one another or gained repressive function. These negative synergies convert quantitative imbalances in TF expression into binary activity patterns. We exploit this principle to automatically create enhancers with specificity to user-defined combinations of hematopoietic progenitor cell states from scratch.
Darwin, nós temos um problema sério: a busca por recuperar (alguma forma de) teleologia continua...
sábado, maio 31, 2025
The Quarterly Review of Biology
Agency in the Evolutionary Transition to Multicellularity
Stuart A. Newman, Mariana Benítez, Ramray Bhat, Tilmann Glimm, K. Vijay Kumar, Vidyanand Nanjundiah, Daniel J. Nicholson, and Sahotra Sarkar
Abstract
This review explores agency, behavior intrinsic to an organism and initiated by it, as it relates to the development of multicellular organisms and its evolution. We ask how agential behaviors contribute to and change concomitantly with evolutionary transitions from unicellularity to multicellularity, including evolution of animals from their closest unicellular antecedents. We consider the relation of organizational properties to the agency of multicellular organisms and conclude, surprisingly, that it is not as strict as it is for individual cells. The main reasons are previously unacknowledged morphogenetic inherencies of multicellular matter and the capacity of development to amplify and partition functionalities of constituent cells. These modalities generate novel phenotypic enablements that enhance the scope of agential behavior. We discuss experimental approaches to distinguish between agency and evolved, stereotypical behaviors of organisms, including purposeful actions. We argue that evolved complexities of animal development make it unsuitable for exploring single-cell-to-multicellular transformations in agency experimentally. We focus our attention instead on agency in the life cycles of social bacteria and amoebae, and in the transitions between multicellular and unicellular states in cancer. Finally, we discuss mathematical representations of incompletely specified dynamical systems and how they may be used to characterize biological autonomy and agency.
FREE PDF: The Quarterly Review of Biology
Darwin, a sua teoria da evolução é tipo uma cidade "Potemkin"?
terça-feira, abril 15, 2025
Is Darwinism a ‘Potemkin’ Theory of Evolution?
The colossal mismatch between life’s observed sophistication and Darwin’s crude mechanism has scientists searching for something more plausible.
by Michael Behe
April 13, 2025, 10:20 PM
Headlines about evolution can give you whiplash. “Was Darwin Wrong?” teased a cover of National Geographic in 2004. But turn the page and you’re conked over the head: “NO. The evidence for Evolution is overwhelming”! Now jump to a recent book review in the journal Nature: “A new vision for how evolution works is long overdue…. [it] challenges a dearly held orthodoxy among evolutionary biologists.”
How could the evidence have been overwhelming but now orthodoxy is being challenged?
The short answer is that Darwin’s theory is a mishmash of multiple logically separate ideas. To avoid confusion, those independent notions have to be teased apart. A couple of strands of Darwinian thought are pretty well supported. Others are close to hopeless. Depending on whether a particular magazine article wants to lecture the rubes that science is in charge, or whip up excitement that something new might be in the air, it emphasizes one or the other.
+++++
Read more here: The American Spectator
Models and Theories - A Philosophical Inquiry, Roman Frigg - Open Access book
sábado, abril 12, 2025
Models and Theories - A Philosophical Inquiry
By Roman Frigg
Edition 1st Edition First Published 2022 eBook Published 27 June 2022 Pub. Location London
Imprint Routledge DOI https://doi.org/10.4324/9781003285106
ABSTRACT
Models and theories are of central importance in science, and scientists spend substantial amounts of time building, testing, comparing and revising models and theories. It is therefore not surprising that the nature of scientific models and theories has been a widely debated topic within the philosophy of science for many years.
The product of two decades of research, this book provides an accessible yet critical introduction to the debates about models and theories within analytical philosophy of science since the 1920s. Roman Frigg surveys and discusses key topics and questions, including:
What are theories? What are models? And how do models and theories relate to each other?
The linguistic view of theories (also known as the syntactic view of theories), covering different articulations of the view, its use of models, the theory-observation divide and the theory-ladenness of observation, and the meaning of theoretical terms.
The model-theoretical view of theories (also known as the semantic view of theories), covering its analysis of the model-world relationship, the internal structure of a theory, and the ontology of models.
Scientific representation, discussing analogy, idealisation and different accounts of representation.
Modelling in scientific practice, examining how models relate to theories and what models are, classifying different kinds of models, and investigating how robustness analysis, perspectivism, and approaches committed to uncertainty-management deal with multi-model situations.
Models and Theories is the first comprehensive book-length treatment of the topic, making it essential reading for advanced undergraduates, researchers, and professional philosophers working in philosophy of science and philosophy of technology. It will also be of interest to philosophically minded readers working in physics, computer sciences and STEM fields more broadly.
FREE PDF GRATIS: Routledge Open Access
Darwin, por que procurar por design se o design na natureza é ilusão?
terça-feira, março 25, 2025
Towards negative carbon footprint: carbon sequestration enabled manufacturing of coral-inspired tough structural composites
Haoxiang Deng, Haixu Du, Ketian Li, Yanchu Zhang, Kyung Hoon Lee, Botong Zheng & Qiming Wang
npj Advanced Manufacturing
Abstract
The increasing impacts of global warming necessitate effective mitigation strategies, with carbon sequestration emerging as a viable mid-term solution. Traditional methods focus on storing CO2 or converting it into liquid substances. However, natural processes like those found in corals demonstrate superior capabilities by transforming CO2 into robust, load-bearing solids with exceptional mechanical properties. Inspired by coral’s biomineralization, this study introduces an electrochemical manufacturing method that converts CO2 into calcium carbonate minerals around 3D-printed polymer scaffolds. This approach results in mineral-polymer composites characterized by extraordinary mechanical strength and fracture toughness, fire resistance, and crack repairability. These composites also offer structure-programmability and composition-reversibility. The scalable modular assembly of these composites supports the creation of larger-scale, load-bearing meso-structures. This manufacturing paradigm promotes negative carbon footprint practices, paving the way for sustainable engineering solutions and a more environmentally friendly future.
FREE PDF GRATIS: npj Advanced Manufacturing Sup. Info.
Darwin, o neodarwinismo é suficiente? Noble e Wilson dialogam sobre a evolução
Is Neo-Darwinism Enough?: The Noble-Wilson Dialogue on Evolution Kindle Edition
by Denis Noble (Author), David Sloan Wilson (Author), James Barham (Editor) Format: Kindle Edition - Amazon
Is Neo-Darwinism Enough? The Noble-Wilson Dialogue on Evolution
David Sloan Wilson and Denis Noble participate in a dialogue centering around the sufficiency of a neo-Darwinian framework of biology. Does neo-Darwinism provide a reasonably complete and adequate explanation of biological phenomena? Are there any new theoretical breakthroughs that are necessary to support the framework? The point of this dialogue is for both parties to advance the best case for their position, as well as to refute the case of their Interlocutor.
Advanced Praise for Is Neo-Darwinism Enough?
Denis Noble and David Sloan Wilson agree on much of what is wrong with the Neo-Darwinian synthesis — and they agree that a lot of things are wrong — yet they part ways on whether these problems amount to an outright falsification of Darwinism (Noble's view) or a call for radically reorienting Darwinism (Wilson's view). Wherever one stands on this issue, their exchange provides an accessible and reliable guide to the unsettled state of contemporary evolutionary theory.
Steve Fuller, Professor of Social Epistemology, University of Warwick UK
Evolution might be as settled a question as any in science. For several decades, how evolution works was similarly portrayed as settled science: Darwinism was the sole credible mechanism, and there was no alternative. The Darwinian idea no longer sits on that throne. But, what is the alternative? In a fascinating dialogue, Is NeoDarwinism Enough? brings together two prominent scientists, David Sloan Wilson, and Denis Noble, to debate the Yea or Nay of the question. Anyone looking for a respectful and balanced exploration of this often contentious question will come away refreshed, better informed, and more fully equipped to decide for themselves.
J. Scott Turner, Emeritus Professor of Biology, SUNY ESF, Syracuse
Public debate over the adequacy and conceptual foundations of the neo-Darwinian synthesis, when it occurs at all, is too often marred by ideology and politics, and more generative of heat than of light. We need dialogue that is not only frank but also civil, reasonable, and well-informed. To that end, Wilson and Noble have made an important contribution.
Edward Feser, Professor of Philosophy, Pasadena City College
David Sloan Wilson is SUNY Distinguished Professor in the departments of Biology and Anthropology at Binghamton University. His research interests focus on multilevel selection theory, human evolution, and differentiation within species, populations, and individuals. Wilson received his PhD from Michigan State University. He is the author of many books, including Evolution for Everyone: How Darwin’s Theory Can Change the Way We Think About Our Lives (Delta, 2007), Does Altruism Exist?: Culture, Genes, and the Welfare of Others (Yale UP, 2015), and This View of Life: Completing the Darwinian Revolution (Pantheon, 2019).
Denis Noble is Emeritus Professor of Cardiovascular Physiology in the Department of Physiology, Anatomy, and Genetics of the Medical Sciences Division of the University of Oxford, where he held the Burdon Sanderson Chair of Cardiovascular Physiology, 1984–2004. A pioneer in computer modeling of biological organs and systems, Noble received his PhD from University College London. He is the author of several books, including The Music of Life: Biology Beyond Genes (Oxford UP, 2008), Dance to the Tune of Life: Biological Relativity (Cambridge UP, 2017), and Understanding Living Systems, with Raymond Noble (Cambridge UP, 2023).
Qual é o papel do desenvolvimento na evolução?
Origem da vida: os micróbios estabeleceram a base para células complexas?
sábado, março 22, 2025
Microtubules in Asgard archaea
Florian Wollweber1,3 ∙ Jingwei Xu (许靖蔚)1,3 ∙ Rafael I. Ponce-Toledo2,4∙ … ∙ Michal Wieczorek1 ∙ Christa Schleper2 ∙ Martin Pilhofer1,5
Highlights
• Asgard archaea express tubulins related to eukaryotic α/β-tubulin and bacterial BtubA/B
• Asgard tubulins (AtubA/B/B2) assemble into canonical and non-canonical heterodimers
• Asgard tubulin heterodimers polymerize into 5 or 7 protofilament microtubules
• AtubA/B form cytoskeletal structures in Ca. Lokiarchaeum ossiferum
Summary
Microtubules are a hallmark of eukaryotes. Archaeal and bacterial homologs of tubulins typically form homopolymers and non-tubular superstructures. The origin of heterodimeric tubulins assembling into microtubules remains unclear.
Here, we report the discovery of microtubule-forming tubulins in Asgard archaea, the closest known relatives of eukaryotes. These Asgard tubulins (AtubA/B) are closely related to eukaryotic α/β-tubulins and the enigmatic bacterial tubulins BtubA/B. Proteomics of Candidatus Lokiarchaeum ossiferum showed that AtubA/B were highly expressed. Cryoelectron microscopy structures demonstrate that AtubA/B form eukaryote-like heterodimers, which assembled into 5-protofilament bona fide microtubules in vitro. The additional paralog AtubB2 lacks a nucleotide-binding site and competitively displaced AtubB. These AtubA/B2 heterodimers polymerized into 7-protofilament non-canonical microtubules. In a sub-population of Ca. Lokiarchaeum ossiferum cells, cryo-tomography revealed tubular structures, while expansion microscopy identified AtubA/B cytoskeletal assemblies.
Our findings suggest a pre-eukaryotic origin of microtubules and provide a framework for understanding the fundamental principles of microtubule assembly.
FREE PDF GRATIS: Cell
Darwin, nós temos um grave problema epistemológico: sua teoria da evolução não pode ser matematizada!
sábado, março 08, 2025
The Reasonable Ineffectiveness of Mathematics in the Biological Sciences
by Seymour Garte 1,* Perry Marshall 2 and Stuart Kauffman 3
1 Department Pharmacology and Toxicology, School of Pharmacy, Rutgers University, Piscataway, NJ 08854, USA
2 Evolution 2.0, Oak Park, IL 60301, USA
3 The Institute for Systems Biology, Seattle, WA 98109-5263, USA
* Author to whom correspondence should be addressed.
Entropy 2025, 27(3), 280; https://doi.org/10.3390/e27030280
Submission received: 24 December 2024 / Revised: 26 February 2025 / Accepted: 5 March 2025 / Published: 7 March 2025
Abstract
The known laws of nature in the physical sciences are well expressed in the language of mathematics, a fact that caused Eugene Wigner to wonder at the “unreasonable effectiveness” of mathematical concepts to explain physical phenomena. The biological sciences, in contrast, have resisted the formulation of precise mathematical laws that model the complexity of the living world. The limits of mathematics in biology are discussed as stemming from the impossibility of constructing a deterministic “Laplacian” model and the failure of set theory to capture the creative nature of evolutionary processes in the biosphere. Indeed, biology transcends the limits of computation. This leads to a necessity of finding new formalisms to describe biological reality, with or without strictly mathematical approaches. In the former case, mathematical expressions that do not demand numerical equivalence (equations) provide useful information without exact predictions. Examples of approximations without equal signs are given. The ineffectiveness of mathematics in biology is an invitation to expand the limits of science and to see that the creativity of nature transcends mathematical formalism.
Keywords: mathematical laws; set theory; third transition
FREE PDF GRATIS: Entropy
Darwin, a ilusão de design na natureza inspira novo material no esqueleto de vidro de uma esponja do mar
quinta-feira, março 06, 2025
Auxetic behavior and energy absorption characteristics of a lattice structure inspired by deep-sea sponge
Jiaming Ma, Hongru Zhang, Ting-Uei Lee, Hongjia Lu, Yi Min Xie, Ngoc San Ha
Centre for Innovative Structures and Materials, School of Engineering, RMIT University, Melbourne 3001, Australia
Received 16 September 2024, Revised 23 December 2024, Accepted 27 December 2024, Available online 27 December 2024, Version of Record 2 January 2025.
https://doi.org/10.1016/j.compstruct.2024.118835
Image/Imagem:The silica skeleton of a Venus’ flower basket sea sponge (Euplectella aspergillum). Credit: RMIT University
Abstract
Auxetic metamaterials, characterized by their lateral contraction under compression, have seen notable progress in recent years, largely due to advancements in 3D printing technologies. However, their practical application remains constrained by limited design versatility, moderate improvements in negative Poisson’s ratio (NPR), and relatively low structural stiffness. To address these challenges, a bio-inspired lattice structure (BLS) has been developed, drawing inspiration from the skeletal system of deep-sea hexactinellid sponges, renowned for their exceptional energy absorption capabilities, stiffness, and mechanical properties. Although this structure exhibits auxetic behavior, a comprehensive understanding of its mechanical performance, including its auxetic properties, remains incomplete. In this study, we systematically explore the auxetic behavior, stiffness, and energy absorption properties of the BLS through a combination of quasi-static compression experiments and detailed numerical simulations using finite element analysis. The experimental results reveal that the BLS outperforms conventional auxetic structures, such as re-entrant hexagonal honeycombs, in terms of NPR, stiffness, and energy absorption capacity. Furthermore, a parametric study is conducted to evaluate the influence of geometric variations, such as member thickness and spacing, on the mechanical performance of the BLS. These findings demonstrate that the BLS has the potential to pioneer a new class of auxetic materials, offering superior mechanical properties and broad applicability in engineering fields that require enhanced energy absorption and structural stiffness under compressive loading.
Keywords
Metamaterial Negative Poisson’s ratio Bio-inspired structure Auxetic 3D printing
FREE PDF GRATIS: Composite Structures
Se o design é mera ilusão na natureza, por que a engenharia aeroespacial se inspira em ossos de pterossauros?
quarta-feira, fevereiro 19, 2025
Harnessing 3D microarchitecture of pterosaur bone using multi-scale X-ray CT for aerospace material design
Nathan Pili, Tristan J. Lowe, Lee Margetts, Kevin Pickup, William I. Sellers, Emma L. Nicholls, Philip J. Withers & Phillip L. Manning
Scientific Reports volume 15, Article number: 5719 (2025)
Image/Imagem: Nathan Pili, The University of Manchester.
Abstract
Pterosaurs were the largest animals to have achieved powered flight in the history of life on Earth, possessing wingspans akin to some modern light aircraft. Vertebrate fossils have shown their potential to retain information on the chemical, physical, and mechanical properties of precursor bone. However, the fossil record is not a traditional source of inspiration for engineers to create palaeo-bioinspired designs. To explore its potential, this study has imaged the three-dimensional porosity of pterosaur bone intending to inspire and improve the mechanical properties of aerospace materials. Historically, two-dimensional histological analysis has resolved fine-scale structures in fossilised bone, which damages the sample. By applying advanced X-ray imaging techniques in this study (using Image Quality Indicators) we show it is possible to non-destructively resolve/verify the microarchitecture of pterosaur bone not previously seen in three dimensions. Pterosaur bone porosity has helped map the macroscopic stresses of this biomaterial but ultimately presents an opportunity to inspire advanced manufactured materials. This microarchitecture of bone offers a unique geometry where self-healing materials with internal monitoring systems can be developed. The iterative process of Darwinian natural selection has evolved multiple engineering solutions that can be reverse engineered to solve challenges facing industry in the 21st Century.
FREE PDF GRATIS: Scientific Reports Sup. Info.
Darwin, nós não temos mais problema algum, pois a evolução se tornou muito boa em evoluir a si mesma.
terça-feira, fevereiro 18, 2025
Evolution takes multiple paths to evolvability when facing environmental change
Bhaskar Kumawat, Alexander Lalejini, Monica M. Acosta, and Luis Zaman
Authors Info & Affiliations
Edited by Paul Turner, Yale University, New Haven, CT; received July 14, 2024; accepted November 27, 2024
December 31, 2024
122 (1) e2413930121
https://doi.org/10.1073/pnas.2413930121
Significance
That all the diversity of life constitutes what Erasmus Darwin called “a single living filament”—an unbroken chain of descent from the last universal common ancestor—is evidence of life’s fundamental adaptability. However, the evolutionary processes that shape this ability to adapt (evolvability) remain elusive because of the required resolution and timespan of observations. Using evolving, self-replicating computer programs, we find that multiple pathways to increased evolvability emerge concurrently and distinctly aid adaptation. One pathway (evolved mutational landscapes) allows rapid adaptation to previously seen environments, while the other (higher mutation rates) allows rapid adaptation to entirely new environments. This multifaceted picture of evolvability helps us understand how organisms deal with ever-changing conditions and relentlessly explore nature’s opportunities for innovation.
Abstract
Life at all scales is surprisingly effective at exploiting new opportunities, as demonstrated by the rapid emergence of antimicrobial resistance and novel pathogens. How populations acquire this level of evolvability and the various ways it aids survival are major open questions with direct implications for human health. Here, we use digital evolution to show that changing environments facilitate the simultaneous evolution of high mutation rates and a distribution of mutational effects skewed toward beneficial phenotypes. The evolved mutational neighborhoods allow rapid adaptation to previously encountered environments, whereas higher mutation rates aid adaptation to completely new environmental conditions. By precisely tracking evolving lineages and the phenotypes of their mutants, we show that evolving populations localize on phenotypic boundaries between distinct regions of genotype space. Our results demonstrate how evolution shapes multiple determinants of evolvability concurrently, fine-tuning a population’s adaptive responses to unpredictable or recurrent environmental shifts.
FREE PDF GRATIS: PNAS
Darwin, nós temos um problema muito sério: não existe uma história verdadeira da vida - discordância de dados
quinta-feira, fevereiro 06, 2025
Biology’s Einstein Moment: Specifying Lineal Frames of Reference and Rejecting Absolute Biological History
Thematic Issue Article
Open access Published: 03 February 2025
Matthew H. Haber
This is the only figure in On the Origin of Species (Darwin 1859). Darwin refers to it numerous times and for different levels of lineage. His presentation introduces a new units question, namely, what are the units of divergence and diversification in an evolutionary system
Abstract
We are currently in the midst of what I call biology’s Einstein moment. This is the rejection of absolute biological history, the idea that there is an invariant, privileged biological history against which other histories are measured or deviate from. Instead, biologists must specify theoretically and empirically motivated frames of lineal reference. This is already informing and advancing biological practice, theory, methods, and more, and is a significant and important feature of contemporary biology. Here I argue that it is worth identifying and naming this shift, and encouraging a deeper and broader embrace of it.
FREE PDF GRATIS: Biological Theory
Excerpt:
"Genealogical discordance like that described above is an important reason that phylogeneticists are moving away from the idea of an absolute phylogeny.”
WOW! Moving away from the idea of an absolute phylogeny? What is the true history of life?
Evidências de colágeno endógeno em osso fóssil de Edmontosaurus, hein?
segunda-feira, fevereiro 03, 2025
Evidence for Endogenous Collagen in Edmontosaurus Fossil Bone
Lucien Tuinstra, Brian Thomas, Steven Robinson, Krzysztof Pawlak, Gazmend Elezi, Kym Francis Faull, Stephen Taylor*
Abstract
Reports of proteins in fossilized bones have been a subject of controversy in the scientific literature because it is assumed that fossilization results in the destruction of all organic components. In this paper, a novel combination of analytical techniques is used to address this question for an exceptionally well-preserved Edmontosaurus sacrum excavated from the Upper Cretaceous strata of the South Dakota Hell Creek Formation. Cross-polarized light microscopy (XPol) shows birefringence consistent with collagen presence. Tandem LC-MS unambiguously identified, and for the first time quantified, hydroxyproline, a unique collagen-indicator amino acid, in acid-digested samples from the Edmontosaurus. LC-MS/MS bottom-up proteomics shows identical collagen peptide sequences previously identified and reported for another hadrosaur and a T. rex sample.
FREE PDF GRATIS: Analytical Chemistry Sup. Info.
Um olhar afiado sobre a navalha de Ockham
sexta-feira, janeiro 31, 2025
Is Ockham’s razor losing its edge? New perspectives on the principle of model parsimony
Marina Dubova mdubova@santafe.edu, Suyog Chandramouli, Gerd Gigerenzer, +11 , and Sabina J. Sloman
Edited by Wilson Geisler, The University of Texas at Austin, Austin, TX; received March 8, 2024; accepted November 19, 2024
January 27, 2025
122 (5) e2401230121
https://doi.org/10.1073/pnas.2401230121
Abstract
The preference for simple explanations, known as the parsimony principle, has long guided the development of scientific theories, hypotheses, and models. Yet recent years have seen a number of successes in employing highly complex models for scientific inquiry (e.g., for 3D protein folding or climate forecasting). In this paper, we reexamine the parsimony principle in light of these scientific and technological advancements. We review recent developments, including the surprising benefits of modeling with more parameters than data, the increasing appreciation of the context-sensitivity of data and misspecification of scientific models, and the development of new modeling tools. By integrating these insights, we reassess the utility of parsimony as a proxy for desirable model traits, such as predictive accuracy, interpretability, effectiveness in guiding new research, and resource efficiency. We conclude that more complex models are sometimes essential for scientific progress, and discuss the ways in which parsimony and complexity can play complementary roles in scientific modeling practice.
FREE PDF GRATIS: PNAS
Ouçam! Ouçam! Os pesquisadores descobrem novas complexidades na audição humana: mero acaso, fortuita necessidade ou design inteligente?
terça-feira, janeiro 28, 2025
Hair Cells in the Cochlea Must Tune Resonant Modes to the Edge of Instability without Destabilizing Collective Modes
Asheesh S. Momi1,2, Michael C. Abbott1,2, Julian Rubinfien1,2, Benjamin B. Machta1,2,*, and Isabella R. Graf1,2,3,†
PRX Life 3, 013001 – Published 2 January, 2025
DOI: https://doi.org/10.1103/PRXLife.3.013001
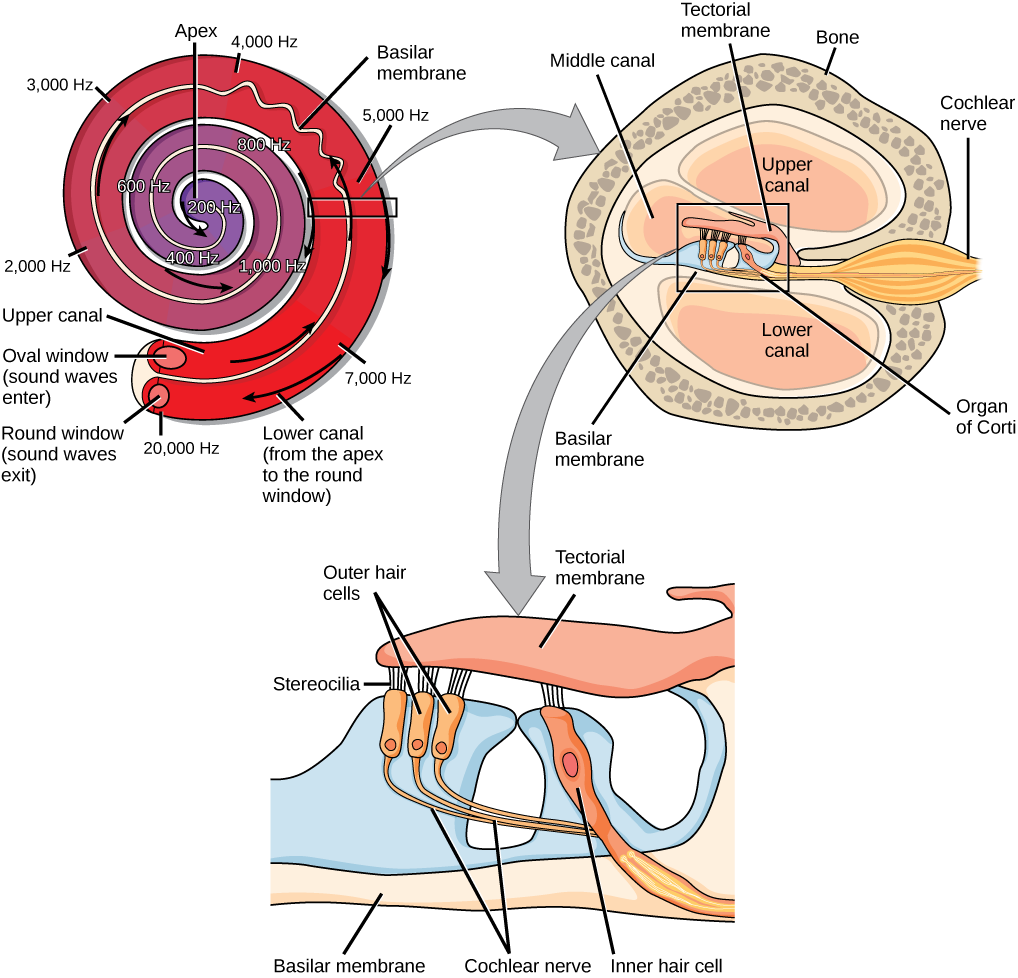
Abstract
Sound produces surface waves along the cochlea's basilar membrane. To achieve the ear's astonishing frequency resolution and sensitivity to faint sounds, dissipation in the cochlea must be canceled via active processes in hair cells, effectively bringing the cochlea to the edge of instability. But how can the cochlea be globally tuned to the edge of instability with only local feedback? To address this question, we use a discretized version of a standard model of basilar membrane dynamics but with an explicit contribution from active processes in hair cells. Surprisingly, we find the basilar membrane supports two qualitatively distinct sets of modes: a continuum of localized modes and a small number of collective extended modes. Localized modes sharply peak at their resonant position and are largely uncoupled. As a result, they can be amplified almost independently from each other by local hair cells via feedback reminiscent of self-organized criticality. However, this amplification can destabilize the collective extended modes; avoiding such instabilities places limits on possible molecular mechanisms for active feedback in hair cells. Our work illuminates how and under what conditions individual hair cells can collectively create a critical cochlea.
FREE PDF GRATIS: PRX LIFE 3
Agência biológica: um conceito sem programa de pesquisa, por que, hein?
terça-feira, janeiro 21, 2025
Biological agency: a concept without a research program
James DiFrisco, Richard Gawne
Journal of Evolutionary Biology, voae153, https://doi.org/10.1093/jeb/voae153
Published: 10 December 2024
Abstract
This paper evaluates recent work purporting to show that the “agency” of organisms is an important phenomenon for evolutionary biology to study. Biological agency is understood as the capacity for goal-directed, self-determining activity—a capacity that is present in all organisms irrespective of their complexity and whether or not they have a nervous system. Proponents of the “agency perspective” on biological systems have claimed that agency is not explainable by physiological or developmental mechanisms, or by adaptation via natural selection. We show that this idea is theoretically unsound and unsupported by current biology. There is no empirical evidence that the agency perspective has the potential to advance experimental research in the life sciences. Instead, the phenomena that the agency perspective purports to make sense of are better explained using the well-established idea that complex multiscale feedback mechanisms evolve through natural selection.
FREE PDF GRATIS: Journal of Evolutionary Biology
Como o darwinismo se esquiva do iceberg
segunda-feira, janeiro 13, 2025
Como o darwinismo se esquiva do iceberg
Neil Thomas
10 de janeiro de 2025 7:43 da manhã
O engenheiro eletrônico escocês Guy Douglas, em seu recém-publicado Evolution's Iceberg, afirma que "as incríveis descobertas no início do final do século 20 e especialmente no século 21, juntamente com as camadas hierárquicas dos sistemas de controle regulatório dentro das células vivas, tornaram-se o 'Iceberg' da Evolution". 1 Sua imagem náutica é apresentada como uma analogia para ligar o destino do Titanic e o suposto "naufrágio" das noções darwinianas tradicionais ocasionadas pelo progresso da biologia molecular no último meio século ou mais. Para ter certeza, a metáfora sobre o darwinismo ser irreparavelmente "violado abaixo da linha d'água" é implicitamente apoiada por cientistas como Micheal Denton, Michael Behe, Douglas Axe e outros agora numerosos demais para citar individualmente. Mas o trabalho dele e o de muitos outros cientistas são realmente o perigo para o paradigma darwiniano que Douglas assume (ou espera?) ser o caso? Eu tenho minhas dúvidas.
Iceberg? Que iceberg?
Se a razão fosse o único árbitro envolvido na discussão, a metáfora do iceberg de Douglas seria totalmente apropriada, mas seu prognóstico deve permanecer problemático pela simples razão de que o HMS Darwin por mais de um século teve permissão para navegar serenamente como se estivesse sobre um lago oceânico com quase nenhum arranhão para frente, para trás ou para o centro. Ignorar esse fato é ignorar o elefante na sala e desconsiderar o fato de que não estamos lidando com ciência empírica aqui (ou mesmo lógica comum), mas com um dogma científico arraigado. Douglas certamente superestima a prontidão dos praticantes da ciência convencional para reexaminar as evidências desapaixonadamente e questionar o que se tornou uma ideia cultural. Para usar o termo adequado de Douglas, a maioria de nós foi aculturada em aceitar o darwinismo, em vez de ter sido educada sobre ele de uma forma devidamente contextualizada (daí o burburinho em muitos sistemas escolares americanos e as disputas legais associadas).
O fato teimoso é que o bom navio Darwin permanece em um estado tão singularmente robusto que uma analogia mais adequada do que o Titanic pode ser com aquele navio de carga de tamanho médio ou "carranca" que desde 1918 está encalhado em um banco de areia a cerca de um quilômetro e meio rio acima da Cachoeira da Ferradura, no meio do rio Niágara (foto no topo). Quando vi aquele navio encalhado em 1992, ele parecia notavelmente bem preservado (além de sua palidez assustadoramente desbotada), mas é claro que funcionalmente era uma mera nave "zumbi" mantida no ar apenas pela camada invisível de rochas logo abaixo da superfície da água.
O espectro da nave atingida que observei naquele dia me parece ser uma imagem mais adequada do que a do Titanic para evocar o status protegido do darwinismo em nossa cultura - um sistema de crenças mantido à tona apenas porque é sustentado metaforicamente por um coro vociferante de partidários que nunca dizem morrer. Portanto, qualquer conversa sobre o eclipse do darwinismo parece prematura e, portanto, para mim, a questão mais relevante e substantiva diz respeito à questão (para reformular a metáfora original de Douglas),
“Por que o Darwinismo NÃO afundou da mesma forma que o Titanic e por que seu naufrágio não parece estar nos planos para breve?”
Essa é uma pergunta que deve ser respondida como um prelúdio indispensável para poder colocar a questão da continuação ou descontinuação do darwinismo na agenda. Infelizmente, é tudo menos fácil de lidar, já que a questão passou a ter mais a ver com religião e secularismo de uma forma ou de outra, do que com a ciência em si (cujo papel no caso às vezes pode parecer alarmantemente tangencial, às vezes quase irrelevante). A ciência do darwinismo (tal como é) muitas vezes atua como uma finta (drible) e como um substituto para debater questões mais profundas de ansiedade cultural. O darwinismo em si, apesar das aparências superficiais, não é o verdadeiro casus belli no conflito, mas simplesmente foi atraído como um ponto de encontro em uma guerra cultural ideológica que, pelo menos em sua forma "quente", só aumentou de intensidade no último meio século ou mais. No que se segue, é claro que darei alguma atenção ao que considero serem os critérios puramente científicos subjacentes ao conflito cultural. Essa tarefa já foi realizada com mais conhecimentos específicos do assunto do que posso reivindicar, e meu foco principal será, portanto, nos aspectos ideológicos igualmente significativos da questão, particularmente no último meio século, uma época que vivi e da qual posso reivindicar algum conhecimento relevante em primeira mão.
Por que, perguntou Annabel Lustig há duas décadas, os argumentos pró e contra Darwin passaram a se assemelhar mais aos antigos procedimentos da Inquisição Espanhola do que ao discurso científico de rotina?2 Em uma tentativa de responder a essa pergunta, começarei com uma breve reprise histórica com a qual nem todos os leitores mais jovens estarão familiarizados, mas que acredito ter desempenhado um papel seminal na consolidação da atitude particularmente intransigente em relação ao darwinismo (a favor ou contra) que testemunhamos desde a década de 1970. Meu foco inicial será o ano de 1963. Essa data é tudo menos arbitrária, pois os eventos daquele ano exerceram uma influência incalculável nas décadas seguintes. Essa influência pode ser facilmente ignorada por aqueles que, por razões compreensíveis, estão mais preocupados com a geração de Woodstock, mais amigável à mídia, e com os inúmeros protestos sociais feitos por aqueles a quem os franceses chamam de les soixante-huitards (a geração de 1968).
1963 - Sementes da Revolução Cultural
Seria um ato de impiedade grosseira não prefaciar o que se segue com o reconhecimento de que o evento mais trágico e literalmente devastador daquele ano ocorreu em 22 de novembro com o assassinato do presidente Kennedy. Nada dessa escala trágica aconteceu na Grã-Bretanha. Ainda assim, foi um ano em que uma série de eventos extraordinários entraram em uma conjunção sinérgica, forçando os britânicos a "cair na real" e fazer uma ruptura decisiva com a era sonolenta da década de 1950, quando ideias mais antigas sobre hierarquia, deferência social, costumes sexuais e piedade convencional não foram questionadas.
Culturalmente, o ano foi marcado, inter alia, pelo julgamento de obscenidade (processado sem sucesso) do livro O Amante de Lady Chatterley de D. H. Lawrence (onde um advogado de acusação ridiculamente advertiu que este não era o tipo de livro que "os servos" deveriam ter permissão para ler) e pelo Caso Profumo (que, em resumo, dependia do flerte do Ministro da Guerra John Profumo com algumas garotas de programas). Tornou-se um imbróglio com outras ramificações que não precisam ser detalhadas aqui, mas que finalmente levaram à queda do governo conservador do primeiro-ministro Harold Macmillan. Acho que deve ser lido como um sintoma daqueles tempos sérios que, quando estudante, fiquei genuinamente chocado com o fato de pessoas em posições de poder e influência poderem se comportar "assim"! Pode ser relevante notar de passagem para os leitores "geração do milênio" que o sexo antes do casamento nesta época, embora quase onipresente, era desaprovado publicamente. Não é de admirar que um muito jovem David Frost 3 tenha conseguido fazer sua estreia na TV em um programa semanal satírico (o imperdível Essa Foi a Semana que Passou) porque havia muita hipocrisia e pensamento duplo para satirizar nos assuntos públicos. Foi o tempo pouco antes da era do Poder das Flores trazer consigo um vento decisivo de mudança nas atitudes das pessoas, apropriadamente incorporado na atitude exuberantemente iconoclasta dos Beatles.
Conversa sobre Deus
Em 1963, tantos ícones do establishment estavam sendo ridicularizados e/ou destronados - então por que não o próprio Deus? De fato, em março de 1963, apareceu (como se fosse uma sugestão!) uma pequena brochura escrita pelo então bispo de Woolwich (Grande Londres), John Robinson, intitulada Honesto com Deus. 5 A importância desta publicação foi tal que os outros eventos mencionados acima parecem, em retrospecto, assumir o status de meros "atos de aquecimento". Pois aqui, em termos de honestidade dolorosa (muitos alegaram "heresia"), o ex-professor de teologia de Cambridge nos disse que devemos abjurar a noção de um Deus pessoal "lá fora" junto com concepções espaciais ingênuas de um "universo de três andares" tradicional. Em vez disso, devemos buscar sinceramente o divino no "fundamento do nosso ser", isto é, dentro de nossa própria psique ou alma: Deus está em nós. Tal como aconteceu com seu colega de Cambridge, Don Cupitt, um pouco mais tarde, ficou claro que Robinson estava abandonando o teísmo objetivo. 6 Essa visão "não-realista" definia "Deus" simplesmente como amor (ágape - devoção altruísta ao bem-estar dos outros) - um ideal espiritual orientador que nos impulsiona de dentro. O ponto principal dessa maneira de pensar é que não poderia mais haver nenhum referente ou pessoa genuinamente transcendente a quem as pessoas pudessem apelar na oração - nenhuma dialética humana/divina. Isso, desnecessário dizer, foi considerado compreensivelmente angustiante por muitos.
Teísmo ou agnosticismo?
A linha divisória entre essa posição e a do humanismo secular parecia tão tênue que era praticamente inexistente e é notável que no prefácio de seu volume, Robinson escreve:
Não raramente, ao assistir ou ouvir uma discussão transmitida entre um cristão e um humanista [secular], me pego percebendo que a maioria das minhas simpatias está do lado do humanista. 7
Pode-se até suspeitar de uma atitude de agnosticismo piedoso por parte do bispo. Em seu relato mais recente do agnosticismo, Robin Le Poidevin afirma que não é apropriado sujeitar proposições religiosas a padrões verificacionistas estritos porque as declarações religiosas são essencialmente julgamentos de valor empregados para expressar um compromisso com um esquema específico de valores e normas morais:
"Embora as sentenças da teologia ou da religião sejam de fato afirmações, elas estão em código. Eles não dizem o que parecem dizer. Eles parecem ser sobre um ser transcendente, o Criador onisciente e todo-poderoso do mundo. O que eles realmente tratam é algo bem diferente: nós, nossos ideais e aspirações, nossa capacidade de amor altruísta e assim por diante." 8
Tal sentimento não parece muito distante da atitude adotada por John Robinson. De fato, o teólogo Alister McGrath foi mais longe ao alegar que Robinson evitou culposamente quaisquer ideias tradicionais de metafísica e concluiu que seu "não-realismo cristão" era, por simples definição, ateísta. 9
O impacto de Honesto com Deus
Um volume complementar de Honest com God foi lançado às pressas pela SCM Press no final de 1963 contendo respostas de clérigos, uma seção transversal de cartas de leitores enviadas a jornais nacionais, algumas resenhas e ensaios de profissionais e um longo adendo do próprio Robinson (pp. 232-279). Um dos revisores, C. S. Lewis, começa sua resposta com uma declaração um tanto surpreendente:
"O bispo de Woolwich perturbará a maioria de nós, leigos cristãos, menos do que ele espera. Há muito abandonamos a crença em um Deus que se senta em um trono em um céu localizado. Chamamos essa crença de antropomorfismo, e ela foi oficialmente condenada antes de nosso tempo." 10
No entanto, Honesto com Deus, desde o momento de sua criação, foi concebido como um livro de bolso para o mercado de massa com um preço de meros cinco xelins (havia 20 xelins por libra), cujo propósito declarado era alcançar o máximo alcance ao maior número de britânicos, congregantes religiosos ou não. Certamente conseguiu isso vendendo mais de um milhão de cópias, e esse número não inclui sua muito discutida serialização em um jornal nacional antes da publicação formal.
No entanto, Honesto com Deus, desde o momento de sua criação, foi concebido como um livro de bolso para o mercado de massa com um preço de meros cinco xelins (havia 20 xelins por libra), cujo propósito declarado era alcançar o máximo alcance ao maior número de britânicos, congregantes religiosos ou não. Certamente conseguiu isso vendendo mais de um milhão de cópias, e esse número não inclui sua muito discutida serialização em um jornal nacional antes da publicação formal.
Como Peter J. Gomes, professor de Moral Cristã em Harvard, apontou 40 anos depois, Robinson estava na verdade dando a seus leitores um extenso tutorial sobre as visões liberais e "desmitologizantes" de teólogos alemães como Rudolf Bultmann, Paul Tillich e Dietrich Bonhoeffer - para não mencionar Ludwig Feuerbach, que já em 1845 havia definido Deus como a "projeção" da consciência moral humana. Gomes apontou que, nesse sentido restrito, Robinson estava transmitindo "conversa de balcão, um discurso entre os conhecedores", mas acrescenta o ponto importante de que Robinson "não percebeu a extensão do analfabetismo teológico da população em geral". 11
E isso provou ser o cerne da questão, pois, a julgar pelas cartas dos leitores reproduzidas no volume Honest to God Debate (pp. 48-81), os esforços de Robinson para apresentar ao público britânico o pensamento teológico avançado se mostraram contraproducentes. O colaborador jornalístico do Sunday Telegraph, T. E. Utley, expressou a decepção e o sentimento de traição de muitas pessoas nos termos mais contundentes e eloqüentes ao explicar como Robinson estava tentando em vão fazer a quadratura de um círculo não quadrado,
"O bispo, é claro, diz que não está tentando destronar Deus, mas redefini-lo de uma maneira aceitável para aqueles que não adotam as premissas da religião cristã ... O propósito declarado do exercício é tornar a religião aceitável para os irreligiosos, provar que é possível ser na realidade um cristão sem acreditar nos ensinamentos da Igreja e aceitar Deus sem usar a palavra ..."
Uma coisa é reafirmar as verdades eternas da religião na linguagem contemporânea [uma referência ao recente aparecimento da Nova Bíblia Inglesa em inglês modernizado no início dos anos sessenta] e outra bem diferente é repudiar expressamente as doutrinas fundamentais que foram cridas por aqueles que aprenderam o cristianismo dos lábios de Cristo .... No mínimo, Robinson parece estar violando os princípios do comércio honesto ao tentar vender como cristã uma mercadoria que não tem relação com o significado histórico e aceito dessa palavra. 12
Utley falou por muitos homens e mulheres comuns que, quando forçados a considerar criticamente a importância de formulários antigos queridos, saíram confusos e desapontados. A desconstrução consciente de Robinson de sua fé parecia-lhes mais um trabalho de demolição. Até mesmo o ultraliberal Don Cupitt afirmou que "o uso da palavra Deus por Robinson parecia estar em todo lugar". 13
Darwin preenche a lacuna de Deus
Não pode haver dúvida de que o caso Honesto com Deus, que começou pouco mais de uma década antes do primeiro dos ataques ateístas de Richard Dawkins (O Gene Egoísta, 1976), teria sido considerado útil para a causa darwiniana. Se os teólogos estavam "falando de Deus para que não existisse" (uma percepção muito comum refletida nas cartas de alguns leitores reimpressas em The Honest to God Debate 14), então o que, além do puro acaso e da chamada "seleção natural", poderia explicar a natureza da ordem criada? Essa questão também estava em primeiro lugar na mente do crítico cultural Christopher Ryan, que objetou que cortar completamente a metafísica e se referir a Deus apenas como a voz mansa e delicada implorava por uma série de perguntas. Certamente não fez nada para explicar o poder instrumental necessário para trazer a criação/evolução em primeiro lugar:
"Uma vez que, em qualquer esfera, a atividade que pode ter sua origem no ser humano, a presença de um alto grau de inteligibilidade das ações defende que essas ações tiveram sua origem em uma inteligência humana, em vez de terem surgido por puro acaso, assim também em esferas de ação que não podem dever sua origem aos seres humanos, a presença de um alto grau de inteligibilidade defende a existência de um Criador, ou seja, um ser racional com o poder de realizar tais ações padronizadas de forma inteligente." 15
Guy Douglas concorda com o argumento do design inteligente de Ryan. Os supostos erros de cópia conhecidos como mutações genéticas devem, pela lei das médias lógicas, levar não à evolução, mas à involução, um deslizamento para baixo, em vez de para cima, do metafórico Monte Improvável de Dawkins. A "auto-organização" recentemente muito debatida, continua ele, pode ser capaz de produzir formas simples de ordem (como flocos de neve), mas não a complexidade especificada necessária para produzir a função biológica. 16 Até o próprio Darwin, ao se referir ao suposto "escrutínio diário e a cada hora" da seleção natural, onde cada melhoria supostamente se fixou no lugar por algum fenômeno de catraca (não especificado), estava, na visão do presente escritor, apenas usando terminologia perifrástica apontando para uma agência divina última. Pois o acaso deixado por conta própria pode claramente produzir apenas aquele caos disfuncional às vezes chamado de entropia.
Aqueles que estão atentos à impossibilidade racional de um poder autônomo e autoatuante de criação e evolução terão ficado pouco perturbados com as noções nebulosas de Robinson, mas pode haver pouca dúvida de que os leitores menos inclinados a pensar sobre as coisas podem não ter notado o fracasso do Bispo Robinson em explicar a natureza e as modalidades da própria Criação. Tais leitores podem ter ficado satisfeitos com a alegação de Lucrécio de que "todo o meio aconteceu por acaso" - uma noção antes considerada uma não-explicação sem valor, mas que agora se tornou a corrente principal.
Falar de seleção natural, evolução de mudança de forma e flutuações quânticas (sustentadas para explicar a criação do universo) são, no final das contas, pouco mais do que uma cortina de fumaça erudita que esconde a ignorância não reconhecida. No entanto, certamente o início da sabedoria deve tomar como ponto de partida o reconhecimento de que existem limites muito definidos para o nosso conhecimento coletivo: a especulação não deve ser confundida com um fato verificável. Um retorno à concepção vitoriana do "mistério dos mistérios" subjacente às nossas vidas pode ser considerado por alguns como um passo retrógrado, mas pelo menos teria a vantagem da honestidade intelectual.
Notas
Guy Douglas, Evolution’s Iceberg: How Molecular Biology Challenges the Theory of Evolution (Lighthouse Christian Publishing: Hochston GA, 2024), p. 75.
Darwinian Heresies, editado por Annabel Lustig, Robert J. Richards, e Michael Ruse (Cambridge: CUP, 2004), Introdução, p. 1.
O mesmo David Frost que mais tarde faria uma entrevista com Richard Nixon após o escândalo de Watergate.
Por ocasião do que foi jocosamente chamado de (re)invasão britânica da América pelo grupo, sua resposta às perguntas do tipo showbiz de um entrevistador americano veio na forma de uma irreverência atrevida de Liverpool que cuidadosamente falhou em se envolver com a substância das perguntas bastante rotineiras do entrevistador.
John A. T. Robinson, Honest to God (Londres: SCM, 1963).
Vide Don Cupitt, Taking Leave of God (Londres: SCM, 1980).
Honest to God, p. 8.
Robin Le Poidevin, Agnosticism: A Very Short Introduction (Oxford: OUP, 2010), citação p. 85.
Alister McGrath, “Jesus for Modern Man: The Historical Significance of John Robinson’s Christology,” in Colin Slee (editor), Honest to God 40 Years On (Londres: SCM, 2004), pp. 111-32.
John Robinson and David L. Edwards, The Honest to God Debate (Londres; SCM, 1963), pp. 91-2, citação p. 91.
Peter J. Gomes. “Honest to God and the Dangerous Ethic” in Honest to God 40 Years On, pp. 70-82, citações pp. 74, 79.
The Honest to God Debate, resenha, pp. 95-97.
“John Robinson and the Language of Faith,” in Honest to God 40 Years On, pp. 37-45, citação p. 38.
See pp. 48-81.
Ryan, “The Language of Theism: Irony and Belief,” in Honest to God 40 Years On, pp. 40-69, citação p. 55.
Evolution’s Iceberg, p. 291.
+++++
Neil Thomas é um reader emérito (um cargo abaixo de professor titular, mas acima de professor sênior) da Universidade de Durham, Inglaterra e membro de longa data da Associação Racionalista Britânica. Ele estudou Estudos Clássicos e Línguas Europeias nas universidades de Oxford, Munique e Cardiff antes de assumir seu cargo na seção alemã da Escola de Línguas e Literaturas Europeias da Universidade de Durham em 1976. Lá, seu ensino envolveu um amplo espectro de especialidades, incluindo filologia germânica, literatura medieval, literatura e filosofia do Iluminismo e história e literatura alemãs modernas. Ele também ensinou módulos sobre o uso propagandista da língua alemã usada tanto pelos nazistas quanto pelos funcionários da antiga República Democrática Alemã. Ele publicou mais de 40 artigos em vários periódicos com revisão por pares e meia dúzia de livros de autoria única, os últimos dos quais foram Lendo o Nibelungenlied (1995), Diu Crone e o Ciclo Arturiano Medieval (2002) e 'Wigalois' de Wirnt von Gravenberg. Intertextualidade e Interpretação (2005). Ele também editou vários volumes, incluindo Mito e seu legado na literatura europeia (1996) e Estudos alemães no milênio (1999). Ele foi o presidente britânico da Sociedade Arturiana Internacional (2002-5) e continua sendo membro de várias sociedades científicas.
Sobre a incompletude inerente das teorias científicas
sexta-feira, janeiro 10, 2025
On the Inherent Incompleteness of Scientific Theories
Philosophy & Ideas
Open access
Published: 24 February 2017
Volume 53, pages 44–100, (2011)
Jolly Mathen
Abstract
We examine the question of whether scientific theories can be complete. For two closely related reasons, we argue that they cannot. The first reason is the inability to determine what are “valid observations”, a result that is based on a self-reference Gödel/Tarski-like argument. The second reason is the existence of “meta-empirical” evidence of the inherent incompleteness of observations. These reasons, along with theoretical incompleteness, are intimately connected to the notion of belief and to theses within the philosophy of science: the Quine-Duhem (and underdetermination) theses and the observational/theoretical distinction failure. Some puzzling aspects of the philosophical theses become clearer in light of these connections. It also follows that there is no absolute measure of the information content of empirical data nor of the entropy of physical systems, and that no complete computer simulation of the natural world is possible. The connections with the mathematical theorems of Gödel and Tarski reveal the existence of other possible connections between scientific and mathematical incompleteness: computational irreducibility, complexity, infinity, arbitrariness, and self-reference. Finally, suggestions are offered of where a more rigorous (or formal) “proof” of scientific incompleteness may be found.
FREE PDF GRATIS: Activitas Nervosa Superior













.%20RMIT%20University.png)










