Did Our Species Evolve in Subdivided Populations across Africa, and Why Does It Matter?
Eleanor M.L. Scerri'Correspondence information about the author Eleanor M.L. ScerriEmail the author Eleanor M.L. Scerri, Mark G. Thomas, Andrea Manica, Philipp Gunz, Jay T. Stock, Chris Stringer, Matt Grove, Huw S. Groucutt, Axel Timmermann, G. Philip Rightmire, Francesco d’Errico, Christian A. Tryon, Nick A. Drake, Alison S. Brooks, Robin W. Dennell, Richard Durbin, Brenna M. Henn, Julia Lee-Thorp, Peter deMenocal, Michael D. Petraglia, Jessica C. Thompson, Aylwyn Scally, Lounès Chikhi
Published Online: July 11, 2018
Publication stage: In Press Corrected Proof
Open access funded by World Health Organization
Highlights
The view that Homo sapiens evolved from a single region/population within Africa has been given primacy in studies of human evolution.
However, developments across multiple fields show that relevant data are no longer consistent with this view
We argue instead that Homo sapiens evolved within a set of interlinked groups living across Africa, whose connectivity changed through time.
Genetic models therefore need to incorporate a more complex view of ancient migration and divergence in Africa.
We summarize this new framework emphasizing population structure, outline how this changes our understanding of human evolution, and identify new research directions.
We challenge the view that our species, Homo sapiens, evolved within a single population and/or region of Africa. The chronology and physical diversity of Pleistocene human fossils suggest that morphologically varied populations pertaining to the H. sapiens clade lived throughout Africa. Similarly, the African archaeological record demonstrates the polycentric origin and persistence of regionally distinct Pleistocene material culture in a variety of paleoecological settings. Genetic studies also indicate that present-day population structure within Africa extends to deep times, paralleling a paleoenvironmental record of shifting and fractured habitable zones. We argue that these fields support an emerging view of a highly structured African prehistory that should be considered in human evolutionary inferences, prompting new interpretations, questions, and interdisciplinary research directions.
A Different View of African Origins
The lineage of Homo sapiens probably originated in Africa at least ∼500 thousand years ago (ka) [1], and the earliest observed morphological manifestations of this clade appeared by ∼300 ka [2]. Early H. sapiens fossils do not demonstrate a simple linear progression towards contemporary human morphology. Instead, putative early H. sapiens remains exhibit remarkable morphological diversity and geographical spread. Together with recent archaeological and genetic lines of evidence, these data are consistent with the view that our species originated and diversified within strongly subdivided (i.e., structured) populations, probably living across Africa, that were connected by sporadic gene flow [1, 3, 4, 5, 6, 7, 8]. This concept of ‘African multiregionalism’ [1] may also include hybridization between H. sapiens and more divergent hominins (see Glossary) living in different regions [1, 9, 10, 11, 12]. Crucially, such population subdivisions may have been shaped and sustained by shifts in ecological boundaries [7, 13, 14], challenging the view that our species was endemic to a single region or habitat, and implying an often underacknowledged complexity to our African origins.
In this paper we examine and synthesize fossil, archaeological, genetic, and paleoenvironmental data to refine our understanding of the time-depth, character, and maintenance of Pleistocene population structure. In doing so, we attempt to separate data from inference to stress that using models of population structure fundamentally changes interpretations of recent human evolution.
The Morphological Diversity and Spread of the Homo sapiens Clade
The constellation of morphological features characterizing H. sapiens is debated. This has strongly impacted on interpretations of recent human origins by variably including or excluding different fossils from interpretative analyses. For example, different morphological criteria and analytical methods have been used to support both a gradual, mosaic-like process of modernization of our species or, conversely, a punctuated speciation (e.g., [1]).
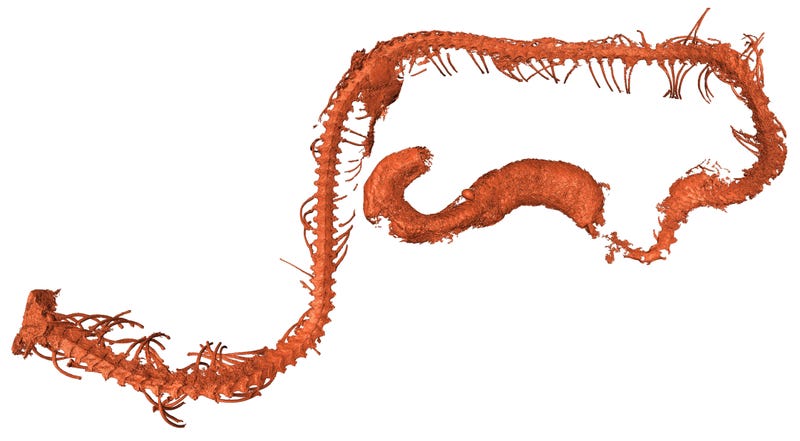
Extant human crania are characterized by a combination of features that distinguish us from our fossil relatives and ancestors, such as a small and gracile face, a chin, and a globular braincase. However, these typical modern human features emerge in a mosaic-like fashion within the H. sapiens clade. The oldest currently recognized members of the H. sapiens clade, from Jebel Irhoud in North Africa, have a facial morphology very similar to extant H. sapiens, as well as endocranial volumes that fall within the contemporary range of variation [2]. However, their braincase shapes are elongated rather than globular, suggesting that distinctive features of brain shape, and possibly brain function, evolved within H. sapiens [2, 5] (Figure 1). Other early H. sapiens fossils from Florisbad in South Africa (∼260 ka), Omo Kibish (∼195 ka) and Herto (∼160 ka), both in Ethiopia, are morphologically diverse [1, 16]. This diversity has led some researchers to propose that fossils such as Jebel Irhoud and Florisbad actually represent a more primitive species called ‘H. helmei’, using the binomen given to the Florisbad partial cranium in 1935 [17, 18]. In a similar vein, the fossil crania from Herto [19], which combine a relatively globular braincase with a robust occipital and large face, were described as the subspecies H. sapiens idaltu because they fall outside the variation of recent humans.
However, we view H. sapiens as an evolving lineage with deep African roots, and therefore prefer to recognize such fossils as part of the diversity shown by early members of the H. sapiens clade. The full suite of cranial features characterizing contemporary humans does not appear until fairly recently, between about ∼100–40 ka [20]. The character and chronology of early H. sapiens fossils, together with their geographic distribution across Africa, suggests that evolution may at times have progressed independently in different regions, in populations that were often semi-isolated for millennia by distance and/or ecological barriers, such as hyperarid regions or tropical forests.
Further insights into the geographic extent and potential habitat diversity of early H. sapiens populations can be gained from more recent forager populations in Africa, which were also strongly structured. For example, Later Stone Age (LSA) human remains highlight both the retention of ‘archaic’ traits and the maintenance of considerable morphological diversity into the terminal Pleistocene [11, 21]. In the Holocene, the skeletal record becomes much richer, but there remains considerable spatial variation in morphology. Variation between populations in different regions and environments of Africa may have been shaped by isolation-by-distance and local environmental adaptations [22, 23, 24, 25, 26]. For example, challenging environments (e.g., deserts, rainforest) and isolation have likely played a significant role in shaping the population structure of Holocene African foragers and isolated hunter-gatherers across the tropics [25, 27].
Ultimately, the processes underlying the emergence of any ‘package’ of derived features diagnostic of early H. sapiens anatomy remain incompletely understood. However, the data do not seem to be consistent with the long-held view that human ancestry is derived predominantly from a single African region hosting a panmictic population. Instead, H. sapiens likely descended from a shifting structured population (i.e., a set of interlinked groups whose connectivity changed through time), each exhibiting different characteristics of anatomical ‘modernity’. The discovery that the primitive-looking H. naledi dates to between ∼335 ka and 236 ka [28], and that the Broken Hill 1 Homo heidelbergensis skull may date to ∼300–125 ka [29], also shows that other hominin species in Africa coexisted with H. sapiens, raising the possibility of African archaic interbreeding. Future research should attempt to determine which features evolved before the appearance of our species and which primarily developed within the evolutionary history of our species. Another key area concerns understanding the extent to which different processes shaped observed changes. For example, the narrowing of the pelvis may reflect different processes including neutral genetic drift, adaptation to ecological variation, and life-history variation.
...