Stephanie A. Carr, Sean P. Jungbluth, Emiley A. Eloe-Fadrosh, Ramunas Stepanauskas, Tanja Woyke, Michael S. Rappé & Beth N. Orcutt
The ISME Journal (2019)
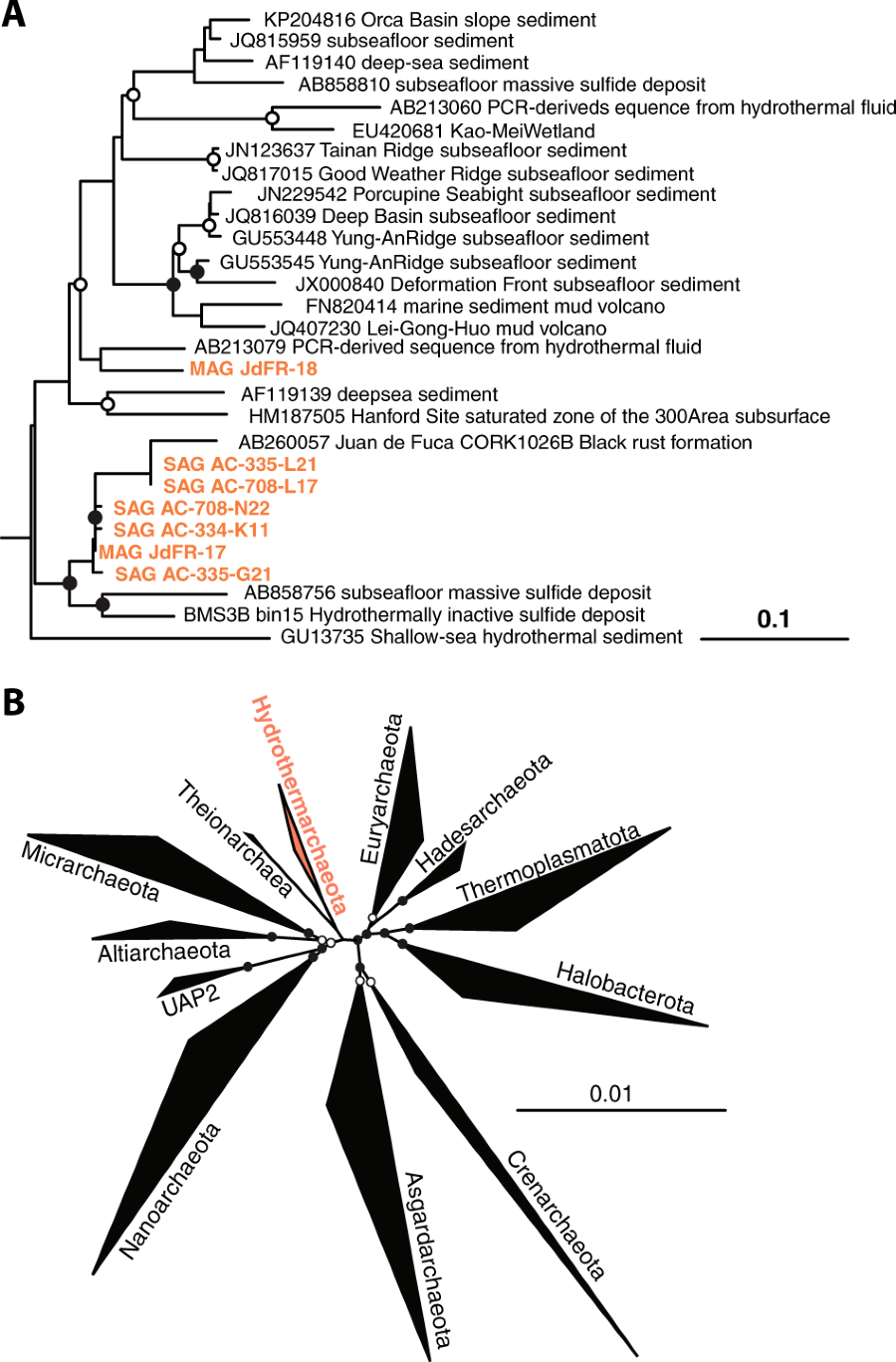
Abstract
The exploration of Earth’s terrestrial subsurface biosphere has led to the discovery of several new archaeal lineages of evolutionary significance. Similarly, the deep subseafloor crustal biosphere also harbors many unique, uncultured archaeal taxa, including those belonging to Candidatus Hydrothermarchaeota, formerly known as Marine Benthic Group-E. Recently, Hydrothermarchaeota was identified as an abundant lineage of Juan de Fuca Ridge flank crustal fluids, suggesting its adaptation to this extreme environment. Through the investigation of single-cell and metagenome-assembled genomes, we provide insight into the lineage’s evolutionary history and metabolic potential. Phylogenomic analysis reveals the Hydrothermarchaeota to be an early-branching archaeal phylum, branching between the superphylum DPANN, Euryarchaeota, and Asgard lineages. Hydrothermarchaeota genomes suggest a potential for dissimilative and assimilative carbon monoxide oxidation (carboxydotrophy), as well as sulfate and nitrate reduction. There is also a prevalence of chemotaxis and motility genes, indicating adaptive strategies for this nutrient-limited fluid-rock environment. These findings provide the first genomic interpretations of the Hydrothermarchaeota phylum and highlight the anoxic, hot, deep marine crustal biosphere as an important habitat for understanding the evolution of early life.
Acknowledgements
The authors thank the AT18-07 cruise party for their assistance including Andrew Fisher as chief scientist; Samuel Hulme, Geoff Wheat, and the late James Cowen for assistance with fluid sampling; Keir Becker for observatory expertise; and Tina Lin, Oliver Hseih, and the late Katrina Edwards for sample handling. We thank the staff of the Single Cell Genomics Center at Bigelow Laboratory for Ocean Sciences for expert handling, sorting, amplification, and sequencing of single cells, the Joint Genome Institute for metagenomic sequencing, Daniel Rogers for mcrA standards, and Bo Barker Jørgensen at Aarhus University for supporting Orcutt’s participation in the cruise. The 2011 cruise was supported by NSF grants MCB-0604014 (lead J. Cowen), OCE-0726887 (lead K. Becker), and OCE-1031808 (lead A. Fisher). SAC was supported by postdoctoral fellowships from the Center for Dark Energy Biosphere Investigations (C-DEBI; NSF award OCE-0939564) and the NSF (OCE-1521614). The efforts of SPJ and MSR were supported by NSF grants MCB-0604014 and OCE-1260723. BNO acknowledges support from C-DEBI, the NASA Astrobiology Institute (NNA13AA92A), and NSF (OCE-1233226). This work was also supported by the grants DEB-1441717 (NSF), OCE-1335810 (NSF) and CSP2013-987 (DOE JGI) to RS. The work conducted by the US Department of Energy Joint Genome Institute (JGI), a US Department of Energy Office of Science User Facility, is supported under contract no. DE-AC02-05CH11231. This is C-DEBI contribution number 451, HIMB contribution number 1747, and SOEST contribution number 10573.
Author information
Affiliations
Bigelow Laboratory for Ocean Sciences, 60 Bigelow Drive, East Boothbay, ME, 04544, USA
Stephanie A. Carr, Ramunas Stepanauskas & Beth N. Orcutt
Hartwick College, Oneonta, NY, USA
Stephanie A. Carr
Center for Dark Energy Biosphere Investigations, University of Southern California, Los Angeles, CA, USA
Sean P. Jungbluth
Department of Energy, Joint Genome Institute, Walnut Creek, CA, USA
Sean P. Jungbluth, Emiley A. Eloe-Fadrosh & Tanja Woyke
Hawaii Institute of Marine Biology, University of Hawaii at Manoa, PO BOX 1346, Kaneohe, HI, 96744, USA
Sean P. Jungbluth & Michael S. Rappé
Conflict of interest
The authors declare that they have no conflict of interest.
Corresponding authors
Correspondence to Michael S. Rappé or Beth N. Orcutt.


