The Role of Orthogonality in Genetic Code Expansion
by Pol Arranz-Gibert 1,2,†,Jaymin R. Patel 1,2,† andFarren J. Isaacs 1,2,*OrcID
1 Department of Molecular, Cellular, and Developmental Biology, Yale University, New Haven, CT 06520, USA
2 Systems Biology Institute, Yale University, West Haven, CT 06516, USA
* Author to whom correspondence should be addressed.
† These authors contributed equally to this work.
Life 2019, 9(3), 58; https://doi.org/10.3390/life9030058
Received: 20 June 2019 / Revised: 1 July 2019 / Accepted: 1 July 2019 / Published: 5 July 2019
(This article belongs to the Special Issue Modelling Life-Like Behavior in Systems Chemistry)
Abstract
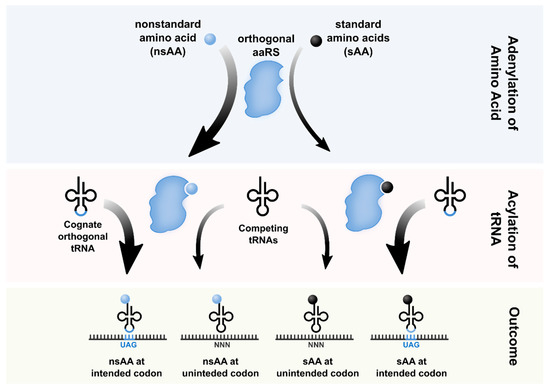
The genetic code defines how information in the genome is translated into protein. Aside from a handful of isolated exceptions, this code is universal. Researchers have developed techniques to artificially expand the genetic code, repurposing codons and translational machinery to incorporate nonstandard amino acids (nsAAs) into proteins. A key challenge for robust genetic code expansion is orthogonality; the engineered machinery used to introduce nsAAs into proteins must co-exist with native translation and gene expression without cross-reactivity or pleiotropy. The issue of orthogonality manifests at several levels, including those of codons, ribosomes, aminoacyl-tRNA synthetases, tRNAs, and elongation factors. In this concept paper, we describe advances in genome recoding, translational engineering and associated challenges rooted in establishing orthogonality needed to expand the genetic code.
Keywords: genetic code expansion; translation; nonstandard amino acids; genome recoding; ribosome engineering; orthogonality; protein engineering
FREE PDF GRATIS: Life


