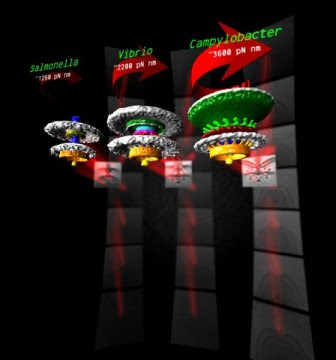
Diverse high-torque bacterial flagellar motors assemble wider stator rings using a conserved protein scaffold
Morgan Beebya,1, Deborah A. Ribardob, Caitlin A. Brennanc,2, Edward G. Rubyc,3, Grant J. Jensend,e, and David R. Hendrixsonb,1
Author Affiliations
Edited by Scott J. Hultgren, Washington University School of Medicine, St. Louis, MO, and approved February 8, 2016 (received for review September 24, 2015)
Abstract Full Text Authors & Info Figures SIMetrics PDF PDF + SI
Source/Fonte: 3 bacterial motors, different torque pN mm - Morgan Beeby - Imperial College London.
Significance
Many bacteria swim using helical propellers, flagella. Intriguingly, different bacteria show different swimming abilities, strikingly illustrated by the abilities of some to bore through viscous fluids (e.g., gastrointestinal mucus) in which others are completely immobilized. We used 3D electron microscopy to show that differences can be explained by the structures of the torque-generating motors: two diverse high-torque motors position additional torque-generating complexes at wider radii from the axial driveshaft than in the model enteric bacteria; this positioning is consistent with the exertion of greater leverage to rotate the flagellum and thus greater torque generation. Intriguingly, these torque-generating complexes are scaffolded at wider radii by a conserved but divergent family of structures, suggesting an ancient origin of reconfiguring torque output.
Abstract
Although it is known that diverse bacterial flagellar motors produce different torques, the mechanism underlying torque variation is unknown. To understand this difference better, we combined genetic analyses with electron cryo-tomography subtomogram averaging to determine in situ structures of flagellar motors that produce different torques, from Campylobacter and Vibrio species. For the first time, to our knowledge, our results unambiguously locate the torque-generating stator complexes and show that diverse high-torque motors use variants of an ancestrally related family of structures to scaffold incorporation of additional stator complexes at wider radii from the axial driveshaft than in the model enteric motor. We identify the protein components of these additional scaffold structures and elucidate their sequential assembly, demonstrating that they are required for stator-complex incorporation. These proteins are widespread, suggesting that different bacteria have tailored torques to specific environments by scaffolding alternative stator placement and number. Our results quantitatively account for different motor torques, complete the assignment of the locations of the major flagellar components, and provide crucial constraints for understanding mechanisms of torque generation and the evolution of multiprotein complexes.
bacterial flagellar motors electron cryo-tomography macromolecular evolution torque Campylobacter
Footnotes
1To whom correspondence may be addressed. Email: mbeeby@imperial.ac.uk or
david.hendrixson@utsouthwestern.edu.
2Present address: Department of Immunology and Infectious Diseases, Harvard T. H. Chan School of Public Health, Boston, MA 02115.
3Present address: Kewalo Marine Laboratory, Pacific Biosciences Research Center, University of Hawaii at Manoa, Honolulu, HI 96813.
Author contributions: M.B., D.A.R., G.J.J., and D.R.H. designed research; M.B. and D.A.R. performed research; M.B., C.A.B., and E.G.R. contributed new reagents/analytic tools; M.B., G.J.J., and D.R.H. analyzed data; and M.B., G.J.J., and D.R.H. wrote the paper.
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The electron cryo-tomography subtomogram average density maps reported in this paper have been deposited in the Electron Microscopy Data Bank (EMD) (accession nos. Salmonella WT: EMD-3154; Vibrio fischeri WT: EMD-3155; Campylobacter jejuni WT: EMD-3150; V. fischeri motB: EMD-3156; C. jejuni motB: EMD-3157; C. jejuni flgQ: EMD-3158; C. jejuni flgP: EMD-3159; C. jejuni pflA: EMD-3160; C. jejuni pflB: EMD-3161; and V. fischeri flgP: EMD-3162).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1518952113/-/DCSupplemental.
Freely available online through the PNAS open access option.
FREE PDF GRATIS: PNAS