Dr. Stephen Meyer Talks about ID 3.0 from Discovery Institute CSC on Vimeo.
Embriões de animais evoluíram antes dos animais
quinta-feira, novembro 28, 2019
The Early Ediacaran Caveasphaera Foreshadows the Evolutionary Origin of Animal-like Embryology
Zongjun Yin6, Kelly Vargas6, John Cunningham, Maoyan Zhu, Federica Marone, Philip Donoghue
Published:November 27, 2019 DOI: https://doi.org/10.1016/j.cub.2019.10.057

• Yin et al. use X-ray tomography to characterize cellular structure and development
• Gastrulation-like cell division, ingression, detachment, and polar aggregation occur
• A holozoan affinity suggests the early evolution of metazoan-like development
Summary
The Ediacaran Weng’an Biota (Doushantuo Formation, 609 Ma old) is a rich microfossil assemblage that preserves biological structure to a subcellular level of fidelity and encompasses a range of developmental stages [1]. However, the animal embryo interpretation of the main components of the biota has been the subject of controversy [2, 3]. Here, we describe the development of Caveasphaera, which varies in morphology from lensoid to a hollow spheroidal cage [4] to a solid spheroid [5] but has largely evaded description and interpretation. Caveasphaera is demonstrably cellular and develops within an envelope by cell division and migration, first defining the spheroidal perimeter via anastomosing cell masses that thicken and ingress as strands of cells that detach and subsequently aggregate in a polar region. Concomitantly, the overall diameter increases as does the volume of the cell mass, but after an initial phase of reductive palinotomy, the volume of individual cells remains the same through development. The process of cell ingression, detachment, and polar aggregation is analogous to gastrulation; together with evidence of functional cell adhesion and development within an envelope, this is suggestive of a holozoan affinity. Parental investment in the embryonic development of Caveasphaera and co-occurring Tianzhushania and Spiralicellula, as well as delayed onset of later development, may reflect an adaptation to the heterogeneous nature of the early Ediacaran nearshore marine environments in which early animals evolved.
FREE PDF GRATIS: Current Biology Sup. Info.
Zongjun Yin6, Kelly Vargas6, John Cunningham, Maoyan Zhu, Federica Marone, Philip Donoghue
Published:November 27, 2019 DOI: https://doi.org/10.1016/j.cub.2019.10.057

Highlights
• Caveasphaera is an enigmatic component of the 609-Ma Weng’an Biota of South China
• Caveasphaera is an enigmatic component of the 609-Ma Weng’an Biota of South China
• Yin et al. use X-ray tomography to characterize cellular structure and development
• Gastrulation-like cell division, ingression, detachment, and polar aggregation occur
• A holozoan affinity suggests the early evolution of metazoan-like development
Summary
The Ediacaran Weng’an Biota (Doushantuo Formation, 609 Ma old) is a rich microfossil assemblage that preserves biological structure to a subcellular level of fidelity and encompasses a range of developmental stages [1]. However, the animal embryo interpretation of the main components of the biota has been the subject of controversy [2, 3]. Here, we describe the development of Caveasphaera, which varies in morphology from lensoid to a hollow spheroidal cage [4] to a solid spheroid [5] but has largely evaded description and interpretation. Caveasphaera is demonstrably cellular and develops within an envelope by cell division and migration, first defining the spheroidal perimeter via anastomosing cell masses that thicken and ingress as strands of cells that detach and subsequently aggregate in a polar region. Concomitantly, the overall diameter increases as does the volume of the cell mass, but after an initial phase of reductive palinotomy, the volume of individual cells remains the same through development. The process of cell ingression, detachment, and polar aggregation is analogous to gastrulation; together with evidence of functional cell adhesion and development within an envelope, this is suggestive of a holozoan affinity. Parental investment in the embryonic development of Caveasphaera and co-occurring Tianzhushania and Spiralicellula, as well as delayed onset of later development, may reflect an adaptation to the heterogeneous nature of the early Ediacaran nearshore marine environments in which early animals evolved.
FREE PDF GRATIS: Current Biology Sup. Info.
Estrutura do gancho flagelar nativo super enrolado como junta universal: mero acaso, fortuita necessidade ou design inteligente?
quarta-feira, novembro 27, 2019
Structure of the native supercoiled flagellar hook as a universal joint
Takayuki Kato, Fumiaki Makino, Tomoko Miyata, Péter Horváth & Keiichi Namba
Nature Communications volume 10, Article number: 5295 (2019)
Structure of native supercoiled hook.
Abstract
The Bacterial flagellar hook is a short supercoiled tubular structure made from a helical assembly of the hook protein FlgE. The hook acts as a universal joint that connects the flagellar basal body and filament, and smoothly transmits torque generated by the rotary motor to the helical filament propeller. In peritrichously flagellated bacteria, the hook allows the filaments to form a bundle behind the cell for swimming, and for the bundle to fall apart for tumbling. Here we report a native supercoiled hook structure at 3.6 Å resolution by cryoEM single particle image analysis of the polyhook. The atomic model built into the three-dimensional (3D) density map reveals the changes in subunit conformation and intersubunit interactions that occur upon compression and extension of the 11 protofilaments during their smoke ring-like rotation. These observations reveal how the hook functions as a dynamic molecular universal joint with high bending flexibility and twisting rigidity.
FREE PDF GRATIS: Nature Communications Sup. Info. Reviewers' File Supplementary Movie 1, 2, 3, 4.
O pipeline de rastreamento de células revela como os circuitos de motores são construídos: mero acaso, fortuita necessidade ou design inteligente?
terça-feira, novembro 26, 2019
Cell Volume 179, ISSUE 2, P355-372.e23, October 03, 2019
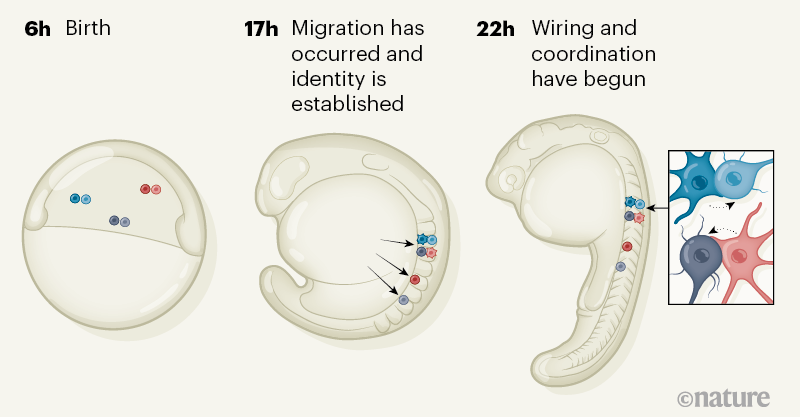
Single-Cell Reconstruction of Emerging Population Activity in an Entire Developing Circuit
Single-Cell Reconstruction of Emerging Population Activity in an Entire Developing Circuit
Yinan Wan, Ziqiang Wei, Loren L. Looger, Minoru Koyama, Shaul Druckmann, Philipp J. Keller
Highlights
• Neurons are tracked from birth to entire circuit at cell-type and functional levels
• Neurogenesis and emergence of coordinated activity is analyzed at a single-cell level
• Motoneurons, active first, form ensembles that synchronize globally, based on size
• Neuron maturation is stereotyped, based on birth time and anatomical origin
Summary
Animal survival requires a functioning nervous system to develop during embryogenesis. Newborn neurons must assemble into circuits producing activity patterns capable of instructing behaviors. Elucidating how this process is coordinated requires new methods that follow maturation and activity of all cells across a developing circuit. We present an imaging method for comprehensively tracking neuron lineages, movements, molecular identities, and activity in the entire developing zebrafish spinal cord, from neurogenesis until the emergence of patterned activity instructing the earliest spontaneous motor behavior. We found that motoneurons are active first and form local patterned ensembles with neighboring neurons. These ensembles merge, synchronize globally after reaching a threshold size, and finally recruit commissural interneurons to orchestrate the left-right alternating patterns important for locomotion in vertebrates. Individual neurons undergo functional maturation stereotypically based on their birth time and anatomical origin. Our study provides a general strategy for reconstructing how functioning circuits emerge during embryogenesis.
FREE PDF GRATIS: Cell
Pare com o treinamento científico que exige "não pergunte"!
sexta-feira, novembro 22, 2019
WORLD VIEW 19 NOVEMBER 2019
Stop the science training that demands ‘don’t ask’
It’s time to trust students to handle doubt and diversity in science, says Jerry Ravetz.
Jerry Ravetz
As a child, I realized that my parents spoke in Yiddish when they didn’t want me to know what they were talking about, so I became aware that some knowledge was intended only for grown-ups — don’t ask. In college, I was taught an elegant theory of chemical combination based on excess electrons going into holes in the orbital shell of a neighbouring atom. But what about diatomic compounds like oxygen gas? Don’t ask; students aren’t ready to know. In physics, I learnt that Newton’s second law of motion is not an empirical, approximate relation such as Boyle’s and Hooke’s laws, and instead has a universal application; but what about the science of statics, in which forces are balanced and there is no acceleration? Don’t ask. Mere students are not worthy of an answer. Yet when I was moonlighting in the social sciences and humanities, I found my questions and opinions were respected, even if only as part of my learning experience.
Observant students will notice that social problems surrounding science are seldom mentioned in official curricula. And now, these pupils are starting to act. They have shamed their seniors into including more diverse contributors as faculty members and role models. Young scholars insolently ask their superiors why they fail to address the extinction crises elucidated by their research. Such subversions are reminiscent of the mass-produced heretical pamphlets circulated by Martin Luther’s supporters at the start of the Protestant Reformation in sixteenth-century Europe. The inherited authoritarian political structures of science education are becoming brittle — but still remain largely unchanged from my own school days.
The philosopher Thomas Kuhn once compared taught science to orthodox theology. A narrow, rigid education does not prepare anyone for the complexities of scientific research, applications and policy. If we discourage students from inquiring into the real nature of scientific truths, or exploring how society shapes the questions that researchers ask, how can we prepare them to maintain public trust in science in our ‘post-truth’ world? Diversity and doubt produce creativity; we must make room for them, and stop funnelling future scientists into narrow specialties that value technique over thought.
In the 1990s, Silvio Funtowicz, a philosopher of science, and I developed the concept of ‘post-normal science’, building on the Kuhnian terms ‘normal’ and ‘revolutionary’ science. It outlines how to use science in a society confronted with high-stakes decisions, where both facts and values are uncertain; it requires drawing on a broad community with broad inquiries. Suppressing questions from budding scientists is sure to suppress promising ideas and solutions.
...
FREE PDF GRATIS: Nature
Sabe aquele DNA "Lixo"? É cheio de informação como os teóricos do Design Inteligente afirmaram!
quarta-feira, novembro 13, 2019
The Genomic Code: A Pervasive Encoding/Molding of Chromatin Structures and a Solution of the “Non‐Coding DNA” Mystery
Giorgio Bernardi
First published: 08 November 2019 https://doi.org/10.1002/bies.201900106

Source/Fonte: Mapping Ignorance
Abstract
Recent investigations have revealed 1) that the isochores of the human genome group into two super‐families characterized by two different long‐range 3D structures, and 2) that these structures, essentially based on the distribution and topology of short sequences, mold primary chromatin domains (and define nucleosome binding). More specifically, GC‐poor, gene‐poor isochores are low‐heterogeneity sequences with oligo‐A spikes that mold the lamina‐associated domains (LADs), whereas GC‐rich, gene‐rich isochores are characterized by single or multiple GC peaks that mold the topologically associating domains (TADs). The formation of these “primary TADs” may be followed by extrusion under the action of cohesin and CTCF. Finally, the genomic code, which is responsible for the pervasive encoding and molding of primary chromatin domains (LADs and primary TADs, namely the “gene spaces”/“spatial compartments”) resolves the longstanding problems of “non‐coding DNA,” “junk DNA,” and “selfish DNA” leading to a new vision of the genome as shaped by DNA sequences.
FREE PDF GRATIS: BioEssays
Fios elétricos vivos conectam-se a tubos sem-fim para maior estabilidade da grade microbiana eletrogênica: mero acaso, fortuita necessidade ou design inteligente?
sexta-feira, novembro 08, 2019
Worm tubes as conduits for the electrogenic microbial grid in marine sediments
Robert C. Aller*, Josephine Y. Aller, Qingzhi Zhu, Christina Heilbrun, Isaac Klingensmith and Aleya Kaushik†
School of Marine and Atmospheric Sciences, Stony Brook University, Stony Brook, NY 11794, USA.
↵*Corresponding author. Email: robert.aller@stonybrook.edu
↵† Present address: NOAA, Global Monitoring Division, Boulder, CO 80305, USA.
Science Advances 17 Jul 2019: Vol. 5, no. 7, eaaw3651
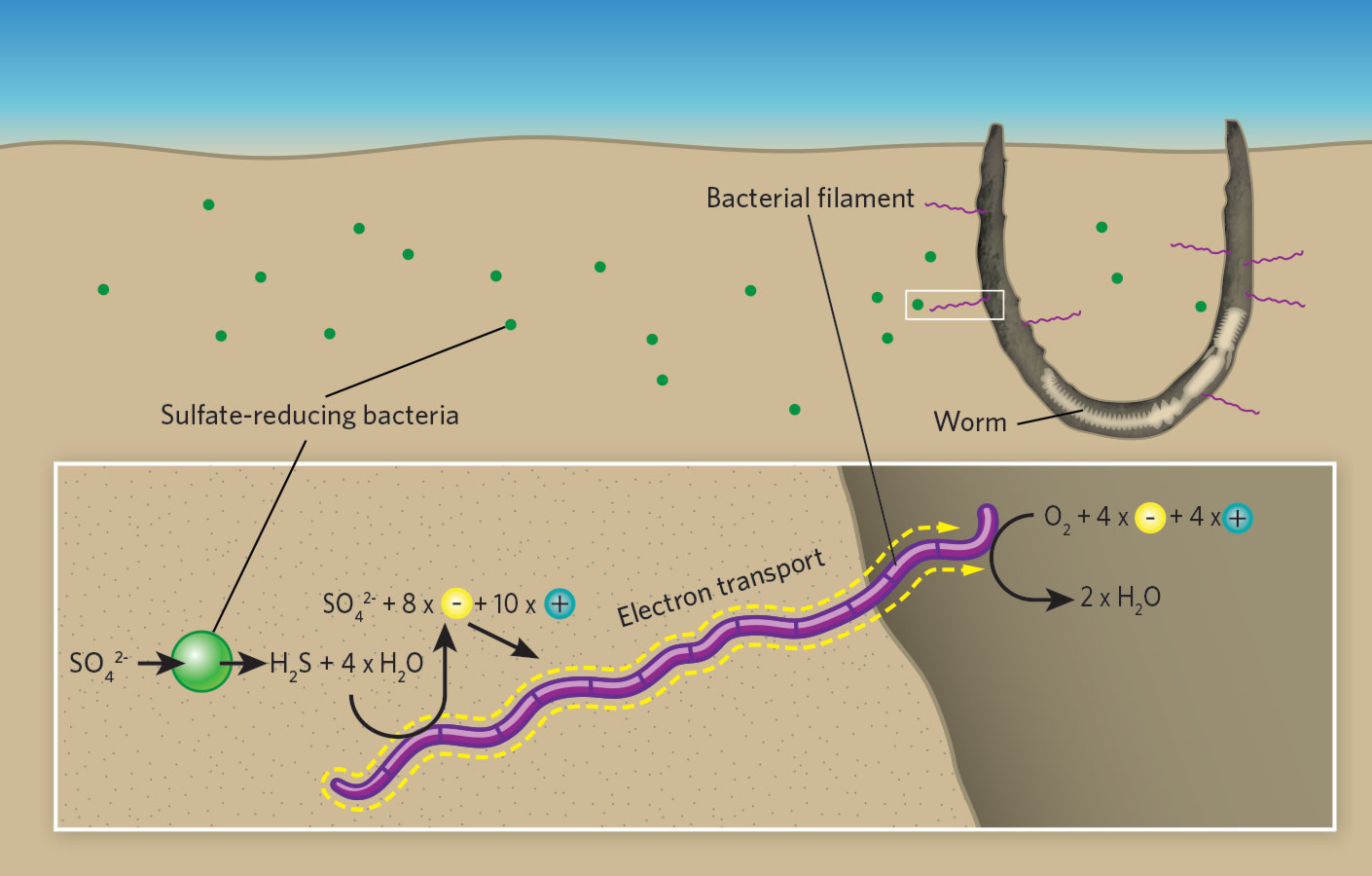
Abstract
Electrogenic cable bacteria can couple spatially separated redox reaction zones in marine sediments using multicellular filaments as electron conductors. Reported as generally absent from disturbed sediments, we have found subsurface cable aggregations associated with tubes of the parchment worm Chaetopterus variopedatus in otherwise intensely bioturbated deposits. Cable bacteria tap into tubes, which act as oxygenated conduits, creating a three-dimensional conducting network extending decimeters into sulfidic deposits. By elevating pH, promoting Mn, Fe-oxide precipitation in tube linings, and depleting S around tubes, they enhance tube preservation and favorable biogeochemical conditions within the tube. The presence of disseminated filaments a few cells in length away from oxygenated interfaces and the reported ability of cable bacteria to use a range of redox reaction couples suggest that these microbes are ubiquitous facultative opportunists and that long filaments are an end-member morphological adaptation to relatively stable redox domains.
FREE PDF GRATIS: Science Advances Sup. Info.
Biologia estrutural celular conforme revelada pela tomografia crioeletrônica: mero acaso, fortuita necessidade ou design inteligente?
quarta-feira, novembro 06, 2019
Cellular structural biology as revealed by cryo-electron tomography
Rossitza N. Irobalieva, Bruno Martins, Ohad Medalia
Journal of Cell Science 2016 129: 469-476; doi: 10.1242/jcs.171967
ABSTRACT
Understanding the function of cellular machines requires a thorough analysis of the structural elements that underline their function. Electron microscopy (EM) has been pivotal in providing information about cellular ultrastructure, as well as macromolecular organization. Biological materials can be physically fixed by vitrification and imaged with cryo-electron tomography (cryo-ET) in a close-to-native condition. Using this technique, one can acquire three-dimensional (3D) information about the macromolecular architecture of cells, depict unique cellular states and reconstruct molecular networks. Technical advances over the last few years, such as improved sample preparation and electron detection methods, have been instrumental in obtaining data with unprecedented structural details. This presents an exciting opportunity to explore the molecular architecture of both individual cells and multicellular organisms at nanometer to subnanometer resolution. In this Commentary, we focus on the recent developments and in situ applications of cryo-ET to cell and structural biology.
FREE PDF GRATIS: Journal of Cell Science
Faleceu Phillip E. Johnson, fundador do Movimento do Design Inteligente
sábado, novembro 02, 2019

Phillip E. Johnson 1940-2019
O papel da ortogonalidade na expansão do código genético: mero acaso, fortuita necessidade ou design inteligente?
quinta-feira, outubro 31, 2019
The Role of Orthogonality in Genetic Code Expansion
by Pol Arranz-Gibert 1,2,†,Jaymin R. Patel 1,2,† andFarren J. Isaacs 1,2,*OrcID
1 Department of Molecular, Cellular, and Developmental Biology, Yale University, New Haven, CT 06520, USA
2 Systems Biology Institute, Yale University, West Haven, CT 06516, USA
* Author to whom correspondence should be addressed.
† These authors contributed equally to this work.
Life 2019, 9(3), 58; https://doi.org/10.3390/life9030058
Received: 20 June 2019 / Revised: 1 July 2019 / Accepted: 1 July 2019 / Published: 5 July 2019
(This article belongs to the Special Issue Modelling Life-Like Behavior in Systems Chemistry)
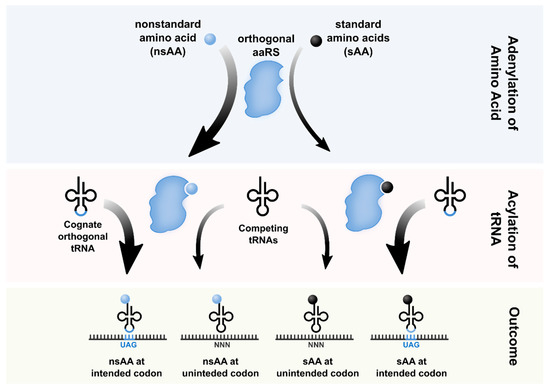
Abstract
The genetic code defines how information in the genome is translated into protein. Aside from a handful of isolated exceptions, this code is universal. Researchers have developed techniques to artificially expand the genetic code, repurposing codons and translational machinery to incorporate nonstandard amino acids (nsAAs) into proteins. A key challenge for robust genetic code expansion is orthogonality; the engineered machinery used to introduce nsAAs into proteins must co-exist with native translation and gene expression without cross-reactivity or pleiotropy. The issue of orthogonality manifests at several levels, including those of codons, ribosomes, aminoacyl-tRNA synthetases, tRNAs, and elongation factors. In this concept paper, we describe advances in genome recoding, translational engineering and associated challenges rooted in establishing orthogonality needed to expand the genetic code.
Keywords: genetic code expansion; translation; nonstandard amino acids; genome recoding; ribosome engineering; orthogonality; protein engineering
FREE PDF GRATIS: Life
Em busca da arqueologia galática
quarta-feira, outubro 30, 2019
In Pursuit of Galactic Archaeology: Astro2020 Science White Paper
Melissa Ness (Columbia/Flatiron), Jonathan Bird (Vanderbilt), Jennifer Johnson (Ohio State University), Gail Zasowski (University of Utah), Juna Kollmeier (Carnegie), Hans-Walter Rix (MPIA), Victor Silva Aguirre (Aarhus), Borja Anguiano (University of Virginia), Sarbani Basu (Yale), Anthony Brown (Leiden), Sven Buder (MPIA), Cristina Chiappini (AIP), Katia Cunha (NOAO), Elena Dongia (University of Wisconsin, Madison), Peter Frinchaboy (TCU), Saskia Hekker (MPI for Solar system research), Jason Hunt (Toronto), Kathryn Johnston (Columbia), Richard Lane (PUC), Sara Lucatello (INAF Osservatorio Astronomico di Padova), Szabolcs Meszaros (ELTE Gothard Astrophysical Observatory)Andres Meza (UDD), Ivan Minchev (AIP), David Nataf (JHU), Marc Pinsonneault (Ohio State University), Adrian M. Price-Whelan (Princeton), Robyn Sanderson (UPenn/Flatiron)Jennifer Sobeck (University of Washington), Keivan Stassun (Vanderbilt), Matthias Steinmetz (AIP), Yuan-Sen Ting (IAS/Princeton/OCIW), Kim Venn (Victoria), Xiangxiang Xue (NAOC)
(Submitted on 11 Jul 2019)
Source/Fonte: The K2 Galactic Archaeology Program
The next decade affords tremendous opportunity to achieve the goals of Galactic archaeology. That is, to reconstruct the evolutionary narrative of the Milky Way, based on the empirical data that describes its current morphological, dynamical, temporal and chemical structures. Here, we describe a path to achieving this goal. The critical observational objective is a Galaxy-scale, contiguous, comprehensive mapping of the disk's phase space, tracing where the majority of the stellar mass resides. An ensemble of recent, ongoing, and imminent surveys are working to deliver such a transformative stellar map. Once this empirical description of the dust-obscured disk is assembled, we will no longer be operationally limited by the observational data. The primary and significant challenge within stellar astronomy and Galactic archaeology will then be in fully utilizing these data. We outline the next-decade framework for obtaining and then realizing the potential of the data to chart the Galactic disk via its stars. One way to support the investment in the massive data assemblage will be to establish a Galactic Archaeology Consortium across the ensemble of stellar missions. This would reflect a long-term commitment to build and support a network of personnel in a dedicated effort to aggregate, engineer, and transform stellar measurements into a comprehensive perspective of our Galaxy.
FREE WHITE PAPER: arxiv
Colocando o "bang" no Big Bang!
terça-feira, outubro 29, 2019
Nonlinear Dynamics of Preheating after Multifield Inflation with Nonminimal Couplings
Rachel Nguyen, Jorinde van de Vis, Evangelos I. Sfakianakis, John T. Giblin, Jr., and David I. Kaiser
Phys. Rev. Lett. 123, 171301 – Published 25 October 2019

Source/Fonte: MIT News
ABSTRACT
We study the postinflation dynamics of multifield models involving nonminimal couplings using lattice simulations to capture significant nonlinear effects like backreaction and rescattering. We measure the effective equation of state and typical timescales for the onset of thermalization, which could affect the usual mapping between predictions for primordial perturbation spectra and measurements of anisotropies in the cosmic microwave background radiation. For large values of the nonminimal coupling constants, we find efficient particle production that gives rise to nearly instantaneous preheating. Moreover, the strong single-field attractor behavior that was previously identified persists until the end of preheating, thereby suppressing typical signatures of multifield models. We therefore find that predictions for primordial observables in this class of models retain a close match to the latest observations.
Revised 10 September 2019Received 10 June 2019
Published by the American Physical Society under the terms of the Creative Commons Attribution 4.0 International license. Further distribution of this work must maintain attribution to the author(s) and the published article’s title, journal citation, and DOI. Funded by SCOAP3.
Published by the American Physical Society
FREE PDF GRATIS: Phys. Rev. Lett.
A ciência não revela a verdade!
Science Does Not Reveal Truth
Paul M. Sutter Contributor
Science
Astrophysicist | Agent to the Stars

If the results can change, is it really true? FREE FOR COMMERCIAL USE (VIA PIXABAY)
What is truth?
It's a pretty popular question, apparently going back several thousand years. And over the centuries and millennia many people have tried to seek out the truth. Or reveal it. Or grapple with the truth. Or wrestle with it. Or at the very least come to terms with it.
And while science is a powerful force in understanding the way the world works, it is not truth.
To draw the distinction, there are facts. There are things we observe about the world around us. We observe the shifting of light from distant galaxies. We observe the mutations over generations in DNA. We can see chemicals combine and interact.
But science itself isn't just a collection of facts about the natural world. That's only part of the story. The bigger part of the story is what we do with those facts. And what we do is try to interpret them and understand them, and fold them into a larger picture that we can use to make predictions about the behavior of those same systems in the natural world.
And all our models and theories are representations and approximations of reality as we see it. That is not truth.
The biggest giveaway is that scientific theories change with time. As we acquire new information or new data, we have to update all of our beliefs. And how can a belief be true if it is subject to change at a moment's notice?
...
Read more: Forbes
Ciência, desprezo e cinismo: cientificismo na nova geração
Science, Scorn, and Cynicism
Scientism in the new generation
By William A. McConnell, Contributing Opinion Writer
William A. McConnell ’21 is a Mathematics concentrator in Adams House. His column appears on alternate Mondays.

Source/Fonte: Thesaurus Plus
It’s an understatement to say that modern science has been the most transformative tool in the human toolkit over the last millennium. From our transportation systems to our digital playthings, we are a society built on the shoulders of science.
We are also a society that has embraced this tool with passion. In elementary school I remember memorizing the scientific method, admiring the genius of Isaac Newton and Marie Curie, and watching with awe my first science experiment, a baking soda and vinegar volcano. STEM education is the stated center of U.S. education policy. News outlets, responding to public demand, now regularly utilize the statistics and studies that their forebears once flippantly labeled nerd-stuff. And wherever we go, we encounter technology, each device quietly advertising to us the power of this tool we call science.
Yet, as (somebody other than, it turns out) Mark Twain famously put it, “If all you have is a hammer, everything looks like a nail.” And thus, amidst this science-oriented backdrop, the doctrine of scientism — “the promotion of science as the best or only objective means by which society should determine normative and epistemological values” — has emerged. It has done so throughout society, but especially in the most significantly exposed segment of the population, the younger generations, the longer educated, and the less religious. We’ve internalized our cultural love of science so that we increasingly tend towards believing that human knowledge is driven by science and science alone, that most questions need to be answered empirically, and that most phenomena can be explained with sufficiently complex physical accounts.
The first problematic symptom of scientism we can identify is the scorn that it often engenders for other modes of thinking. The religious arena best illuminates this scorn. At some point we all probably have seen, or have been, the person who confidently bashes on religion for having no evidence behind it. “Evidence” needs no adjective — for of course it is only evidence of the scientific sort that counts. And thus the ship of scientism leaves behind the religious, with an amused chuckle and a petulant puff of steam. My point here is less to initiate a metaphysical conversation about god/God than it is to illustrate how our scientism often precludes us from even getting to a starting point for such a conversation.
This same attitude possesses conversations that deal with topics like free will, morality, the nature of the mind, and the nature of gender. These questions do not ask for experiments but for reason, for examples and analogies, for theories of the meanings of words, for hypotheticals and counterfactuals, for powerful explanations, and for reflections on our own lived experiences. Yet our capacity and propensity to think along these lines fade as we become more and more convinced that truth in this world can be reduced to science. We become mired in the kind of thinking that says free will and morality cannot exist in an entirely physical universe, and that minds and gender can be reduced to the physical processes underlying them. We skip the humanities for the hard sciences, supposedly the only place where real, productive thinking takes place.
Scientism cuts deeper, however, in the way that it inherently devalues the individual. It denies the capabilities of an individual mind, and the meaning of an individual existence, thus cultivating repressed cynicism about the world.
...
Read more here: The Crimson
Sondando a complexidade: abordagens termodinâmica e mecânica computacional para estudos das origens
quinta-feira, outubro 24, 2019
Research article
Probing complexity: thermodynamics and computational mechanics approaches to origins studies
Stuart J. Bartlett and Patrick Beckett
Published:18 October 2019 https://doi.org/10.1098/rsfs.2019.0058
Abstract
This paper proposes new avenues for origins research that apply modern concepts from stochastic thermodynamics, information thermodynamics and complexity science. Most approaches to the emergence of life prioritize certain compounds, reaction pathways, environments or phenomena. What they all have in common is the objective of reaching a state that is recognizably alive, usually positing the need for an evolutionary process. As with life itself, this correlates with a growth in the complexity of the system over time. Complexity often takes the form of an intuition or a proxy for a phenomenon that defies complete understanding. However, recent progress in several theoretical fields allows the rigorous computation of complexity. We thus propose that measurement and control of the complexity and information content of origins-relevant systems can provide novel insights that are absent in other approaches. Since we have no guarantee that the earliest forms of life (or alien life) used the same materials and processes as extant life, an appeal to complexity and information processing provides a more objective and agnostic approach to the search for life's beginnings. This paper gives an accessible overview of the three relevant branches of modern thermodynamics. These frameworks are not commonly applied in origins studies, but are ideally suited to the analysis of such non-equilibrium systems. We present proposals for the application of these concepts in both theoretical and experimental origins settings.
FREE PDF GRATIS: Interface Focus
The Gene Emergence Project - artigos de David L. Abel com revisão por pares
sexta-feira, outubro 18, 2019
The Gene Emergence Project is one of the programs of The Origin-of-Life Science Foundation, Inc., a 501(c)3 science and education foundation with corporate headquarters near NASA's Goddard Space Flight Center just off the Washington, D. C. Beltway in Greenbelt, MD. 113 Hedgewood Drive, 20770-1610 Fax 301-441-8135
Dr. Abel is a life-origin specialist with scores of peer-reviewed science journal publications. He is the Editor of The First Gene: The Birth of Programming, Messaging and Formal Control.
Dr. Abel is the Director of the Gene Emergence Project of the Origin of Life Science Foundation, Inc. His specialties are Proto-Biocybernetics and Proto-Biosemiotics.
His latest book is Primordial Prescription: the Most Plaguing Problem of Life-Origin Science
Many of Dr. Abel's science journal papers are downloadable from http://lifeorigin.academia.edu/DrDavidLAbel
Most of his peer-reviewed science journal papers also downloadable from http://www.researchgate.net/
The Origin-of-Life Science Foundation should not be confused with "creation science"or "intelligent design" groups. It has no religious affiliations of any kind, nor are we connected in any way with any New Age, Gaia, or "Science and Spirit" groups. The Origin-of-Life Science Foundation, Inc. is a science and education foundation encouraging the pursuit of natural-process explanations and mechanisms within nature. The Foundation's main thrust is to encourage interdisciplinary, multi-institutional research projects by theoretical biophysicists and origin-of-life researchers specifically into the origin of genetic information/instructions/message/recipe in living organisms. By what mechanism did initial genetic code arise in nature? The primary interest of The Gene Emergence Project is to investigate the derivation of functional monomeric sequencing at the rigid covalent-bond level. This must occur prior to any selection for phenotypic fitness.
Fitting with the project's highly interdisciplinary nature, its advisors include biochemists, molecular biologists, biophysicists, information theorists, artificial life and intelligence experts, exo/astrobiologists, mathemeticians, and origin-of-life researchers in many related fields. Please feel free to e-mail or write us opinions, advice, and critiques, particularly of the tentative rules themselves. We are developing as broad and as deep a root system within the scientific community as possible.
The Foundation believes that advisors' personal metaphysical persuasions are none of our business. Science is about "How?" Questions addressing "How?" are about mechanism. As a science foundation, we are interested in models of mechanism consistent with naturally-occuring biochemical phenomena and sound information theory. We welcome as advisors competent scientists from widely respected universities and laboratories around the world whose interest and experience extends to origin-of-life queries.
The Foundation welcomes tax-deductible donations to be used for the promotion of scientific inquiry into the origin of genetic prescriptive information (instruction).
The Fundamental Questions for Life Origin Research -
Why and how would a prebiotic environment value, desire or seek to generate utility?
The peer-reviewed literature referenced in this website provides valuable background information showing why life-origin theorists struggle to generate a model or theory that addresses any of the most important questions of life origin.
An additional book of interest is: Programming of Life, by Johnson, D.E., Big Mac Publishers: Sylacauga, Alabama, 2010; p 127.
Dr. Abel is the Director of the Gene Emergence Project of the Origin of Life Science Foundation, Inc. His specialties are Proto-Biocybernetics and Proto-Biosemiotics.
His latest book is Primordial Prescription: the Most Plaguing Problem of Life-Origin Science
Many of Dr. Abel's science journal papers are downloadable from http://lifeorigin.academia.edu/DrDavidLAbel
Most of his peer-reviewed science journal papers also downloadable from http://www.researchgate.net/
The Origin-of-Life Science Foundation should not be confused with "creation science"or "intelligent design" groups. It has no religious affiliations of any kind, nor are we connected in any way with any New Age, Gaia, or "Science and Spirit" groups. The Origin-of-Life Science Foundation, Inc. is a science and education foundation encouraging the pursuit of natural-process explanations and mechanisms within nature. The Foundation's main thrust is to encourage interdisciplinary, multi-institutional research projects by theoretical biophysicists and origin-of-life researchers specifically into the origin of genetic information/instructions/message/recipe in living organisms. By what mechanism did initial genetic code arise in nature? The primary interest of The Gene Emergence Project is to investigate the derivation of functional monomeric sequencing at the rigid covalent-bond level. This must occur prior to any selection for phenotypic fitness.
Fitting with the project's highly interdisciplinary nature, its advisors include biochemists, molecular biologists, biophysicists, information theorists, artificial life and intelligence experts, exo/astrobiologists, mathemeticians, and origin-of-life researchers in many related fields. Please feel free to e-mail or write us opinions, advice, and critiques, particularly of the tentative rules themselves. We are developing as broad and as deep a root system within the scientific community as possible.
The Foundation believes that advisors' personal metaphysical persuasions are none of our business. Science is about "How?" Questions addressing "How?" are about mechanism. As a science foundation, we are interested in models of mechanism consistent with naturally-occuring biochemical phenomena and sound information theory. We welcome as advisors competent scientists from widely respected universities and laboratories around the world whose interest and experience extends to origin-of-life queries.
The Foundation welcomes tax-deductible donations to be used for the promotion of scientific inquiry into the origin of genetic prescriptive information (instruction).
The Fundamental Questions for Life Origin Research -
How did molecular evolution generate metabolic recipe and instructions using a representational symbol system?
How did prebiotic nature set all of the many configurable switch-settings to integrate so many interdependent circuits?
How did inanimate nature sequence nucleotides to spell instructions to the ribosomes on how to sequence amino acids into correctly folding proteins?
How did nature then code these instructions into Hamming block codes to reduce noise pollution in the Shannon channel?
What programmed the error-detection and error-correcting software that keeps life from quickly deteriorating into non-life?
In short, which of the four known forces of physics organized and prescribed life into existence? Was it gravity? Was it the strong or weak nuclear force? Was it the electromagnetic force? How could any combination of these natural forces or force fields program decision nodes to prescribe future utility?
Can chance and/or necessity program or prescribe sophisticated biofunction?
Life is utterly dependent upon the steering of reaction sequences into biochemical pathways and cycles.
Life pursues the goal of staying alive. All known life is cybernetic, meaning controlled. Life's most prominent attribute is programming and tight regulation at every turn. Yet programming, prescription, control and regulation are all formalisms, not mere physicodynamic interactions. The programming of life is what makes life unique [1-3]
Metabolism First models cannot sustain themselves as perpetual motion machines, even in open environments, without heritable formal instructions needed to circumvent locally and temporarily 2nd Law organizational and useful energy deterioration.
Prescription and programming arise only out of Decision Theory, not Stochastic Theory.
How did prebiotic nature program the first decision nodes? Only Choice-Contingent Causation and Control (CCCC) could possibly program a genome and epigenome.
The peer-reviewed literature referenced in this website provides valuable background information showing why life-origin theorists struggle to generate a model or theory that addresses any of the most important questions of life origin.
An additional book of interest is: Programming of Life, by Johnson, D.E., Big Mac Publishers: Sylacauga, Alabama, 2010; p 127.
As células da mitocôndria funcionam muito parecido com pacote de baterias Tesla: mero acaso, fortuita necessidade ou design inteligente?
quinta-feira, outubro 17, 2019
Individual cristae within the same mitochondrion display different membrane potentials and are functionally independent
Dane M Wolf
Department of Medicine (Endocrinology), Department of Molecular and Medical Pharmacology, David Geffen School of Medicine, University of California, Los Angeles, CA, USA
Graduate Program in Nutrition and Metabolism, Graduate Medical Sciences, Boston University School of Medicine, Boston, MA, USA
Search for more papers by this author
Graduate Program in Nutrition and Metabolism, Graduate Medical Sciences, Boston University School of Medicine, Boston, MA, USA
Search for more papers by this author
Department of Medicine (Endocrinology), Department of Molecular and Medical Pharmacology, David Geffen School of Medicine, University of California, Los Angeles, CA, USA
Institute of Biochemistry and Molecular Biology I, Medical Faculty, Heinrich Heine University Düsseldorf, Düsseldorf, Germany
Institute of Biochemistry and Molecular Biology I, Medical Faculty, Heinrich Heine University Düsseldorf, Düsseldorf, Germany
Department of Pulmonary and Critical Care Medicine, David Geffen School of Medicine, University of California, Los Angeles, CA, USA
Jonsson Comprehensive Cancer Center, David Geffen School of Medicine, University of California, Los Angeles, CA, USA
Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA
Institute of Biochemistry and Molecular Biology I, Medical Faculty, Heinrich Heine University Düsseldorf, Düsseldorf, Germany
Molecular Biology Institute at UCLA, Los Angeles, CA, USA
Department of Biological Chemistry, David Geffen School of Medicine at UCLA, Los Angeles, CA, USA
Department of Pulmonary and Critical Care Medicine, David Geffen School of Medicine, University of California, Los Angeles, CA, USA
Jonsson Comprehensive Cancer Center, David Geffen School of Medicine, University of California, Los Angeles, CA, USA
Corresponding Author
E-mail address: mliesa@mednet.ucla.edu
Department of Medicine (Endocrinology), Department of Molecular and Medical Pharmacology, David Geffen School of Medicine, University of California, Los Angeles, CA, USA
Molecular Biology Institute at UCLA, Los Angeles, CA, USA
Corresponding Author
E-mail address: OShirihai@mednet.ucla.edu
Department of Medicine (Endocrinology), Department of Molecular and Medical Pharmacology, David Geffen School of Medicine, University of California, Los Angeles, CA, USA
Graduate Program in Nutrition and Metabolism, Graduate Medical Sciences, Boston University School of Medicine, Boston, MA, USA
Dane M Wolf
Department of Medicine (Endocrinology), Department of Molecular and Medical Pharmacology, David Geffen School of Medicine, University of California, Los Angeles, CA, USA
Graduate Program in Nutrition and Metabolism, Graduate Medical Sciences, Boston University School of Medicine, Boston, MA, USA
Search for more papers by this author
Graduate Program in Nutrition and Metabolism, Graduate Medical Sciences, Boston University School of Medicine, Boston, MA, USA
Search for more papers by this author
Mayuko Segawa
Department of Medicine (Endocrinology), Department of Molecular and Medical Pharmacology, David Geffen School of Medicine, University of California, Los Angeles, CA, USA
Search for more papers by this author
Search for more papers by this author
Arun Kumar Kondadi
Institute of Biochemistry and Molecular Biology I, Medical Faculty, Heinrich Heine University Düsseldorf, Düsseldorf, Germany
Search for more papers by this author
Search for more papers by this author
Ruchika Anand
Institute of Biochemistry and Molecular Biology I, Medical Faculty, Heinrich Heine University Düsseldorf, Düsseldorf, Germany
Search for more papers by this author
Search for more papers by this author
Sean T Bailey
Department of Pulmonary and Critical Care Medicine, David Geffen School of Medicine, University of California, Los Angeles, CA, USA
Jonsson Comprehensive Cancer Center, David Geffen School of Medicine, University of California, Los Angeles, CA, USA
Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA
Search for more papers by this author
Jonsson Comprehensive Cancer Center, David Geffen School of Medicine, University of California, Los Angeles, CA, USA
Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA
Search for more papers by this author
Andreas S Reichert
Institute of Biochemistry and Molecular Biology I, Medical Faculty, Heinrich Heine University Düsseldorf, Düsseldorf, Germany
Search for more papers by this author
Search for more papers by this author
Alexander M van der Bliek
Molecular Biology Institute at UCLA, Los Angeles, CA, USA
Department of Biological Chemistry, David Geffen School of Medicine at UCLA, Los Angeles, CA, USA
Search for more papers by this author
Department of Biological Chemistry, David Geffen School of Medicine at UCLA, Los Angeles, CA, USA
Search for more papers by this author
David B Shackelford
Department of Pulmonary and Critical Care Medicine, David Geffen School of Medicine, University of California, Los Angeles, CA, USA
Jonsson Comprehensive Cancer Center, David Geffen School of Medicine, University of California, Los Angeles, CA, USA
Search for more papers by this author
Jonsson Comprehensive Cancer Center, David Geffen School of Medicine, University of California, Los Angeles, CA, USA
Search for more papers by this author
Marc Liesa
E-mail address: mliesa@mednet.ucla.edu
Department of Medicine (Endocrinology), Department of Molecular and Medical Pharmacology, David Geffen School of Medicine, University of California, Los Angeles, CA, USA
Molecular Biology Institute at UCLA, Los Angeles, CA, USA
Search for more papers by this author
Corresponding Author
E-mail address: mliesa@mednet.ucla.edu
Department of Medicine (Endocrinology), Department of Molecular and Medical Pharmacology, David Geffen School of Medicine, University of California, Los Angeles, CA, USA
Molecular Biology Institute at UCLA, Los Angeles, CA, USA
Search for more papers by this author
Orian S Shirihai
E-mail address: OShirihai@mednet.ucla.edu
Department of Medicine (Endocrinology), Department of Molecular and Medical Pharmacology, David Geffen School of Medicine, University of California, Los Angeles, CA, USA
Graduate Program in Nutrition and Metabolism, Graduate Medical Sciences, Boston University School of Medicine, Boston, MA, USA
Search for more papers by this author
Corresponding Author
E-mail address: OShirihai@mednet.ucla.edu
Department of Medicine (Endocrinology), Department of Molecular and Medical Pharmacology, David Geffen School of Medicine, University of California, Los Angeles, CA, USA
Graduate Program in Nutrition and Metabolism, Graduate Medical Sciences, Boston University School of Medicine, Boston, MA, USA
Search for more papers by this author
- *Corresponding author. Tel: +1‐310‐206‐7319; E‐mail: mliesa@mednet.ucla.edu
EMBO J (2019)e101056https://doi.org/10.15252/embj.2018101056
Abstract
The mitochondrial membrane potential (ΔΨm) is the main driver of oxidative phosphorylation (OXPHOS). The inner mitochondrial membrane (IMM), consisting of cristae and inner boundary membranes (IBM), is considered to carry a uniform ΔΨm. However, sequestration of OXPHOS components in cristae membranes necessitates a re‐examination of the equipotential representation of the IMM. We developed an approach to monitor ΔΨm at the resolution of individual cristae. We found that the IMM was divided into segments with distinct ΔΨm, corresponding to cristae and IBM. ΔΨm was higher at cristae compared to IBM. Treatment with oligomycin increased, whereas FCCP decreased, ΔΨm heterogeneity along the IMM. Impairment of cristae structure through deletion of MICOS‐complex components or Opa1 diminished this intramitochondrial heterogeneity of ΔΨm. Lastly, we determined that different cristae within the individual mitochondrion can have disparate membrane potentials and that interventions causing acute depolarization may affect some cristae while sparing others. Altogether, our data support a new model in which cristae within the same mitochondrion behave as independent bioenergetic units, preventing the failure of specific cristae from spreading dysfunction to the rest.
FREE PDF GRATIS: EMBO J
Assinar:
Postagens (Atom)