Desafios matemáticos à teoria da evolução - Berlinski, Meyer, e Gelernter
segunda-feira, julho 22, 2019
Origem da vida: tempo esgotado!
sexta-feira, julho 19, 2019
Time Out
James Tour
In 1952, Stanley Miller and Harold Urey derived a number of racemic amino acids from a handful of small molecules. These were electrifying results because they suggested that the methods of synthetic chemistry might finally explain the origins of life. The excitement was justified, but premature.1 Origins of life (OOL) research has, to be sure, become progressively more sophisticated, but its goal—to explain the origins of life—remains as distant today as it was in 1952. This is not surprising. The protocols in use have remained unchanged: buy highly purified chemicals; mix them together in high concentrations and in a specific order under carefully devised laboratory conditions; derive a mixture of compounds; and publish a paper making bold claims about OOL. These protocols are as unrealistic as they are unimproved.
This essay comprises an argument, but it also contains an appeal to the OOL community. The history of science suggests that on occasion what is required for research to flourish is not further research—at least to the extent that further research involves doing the same thing. This is one of those times.
Needed for Life
Four molecules are needed for life: nucleotides, carbohydrates, proteins, and lipids. Nucleotides are composed of a trimeric nucleobase-carbohydrate-phosphate combination, and once polymerized, constitute DNA and RNA. Five different nucleobases comprise the entire alphabet for DNA and RNA. The nucleotides and their subsequent DNA and RNA structures are homochiral, yielding one of two possible enantiomers. Amino acids are most often homochiral. When amino acids are polymerized, they form proteins and enzymes. Proteins and enzymes also display a tertiary homochirality. Lipids are dipolar molecules with a polar water-soluble head and a non-polar water-insoluble tail. They, too, are most often homochiral. Cells use carbohydrates for energy, and carbohydrates, along with proteins, are identification-receptors. Carbohydrates are also homochiral, and their polymeric forms take on tertiary homochiral shapes. OOL researchers have spent a great deal of time trying to make these four classes of molecules, but with scant success.
Constructing the molecules necessary for life from their prebiotic precursors represents one goal of OOL research; putting them together, another. Some of synthetic chemistry is pedestrian, and some ingenious. Fundamental questions remain unaddressed. Claims that these structures could be prepared under prebiotic conditions in high enantiomeric purity using inorganic templates, or any presumed templates, have never been realized. The carbohydrates, amino acids, lipids, and other compounds within each of these classes require specific methods in order to control their regiochemistry and stereochemistry. The differences in reaction rates often require chiral systems acting upon chiral molecules. If this were possible under prebiotic conditions, it is odd that it cannot be replicated by synthetic chemists.
They have, after all, had 67 years to try.
Synthetic Hyperbole
Consider the class of experiments that deal with the assembly of chemicals into what are referred to as protocells—“a self-organized, endogenously ordered, spherical collection of lipids proposed as a stepping-stone to the origin of life.”2 In 2017, a team from the Origins of Life Initiative at Harvard University performed a type of polymerization reaction in water known as the reversible addition–fragmentation chain transfer.3 This reaction type is not seen in nature, and neither are the monomers that figure in the experiment. Still, this is standard chemistry. Polymers are made by a controlled radical polymerization reaction, where two different monomer types are added sequentially to a chain bearing both a hydrophobic and a hydrophilic block. Researchers observed polymeric vesicles forming during polymerization—interesting, but not extraordinary. The vesicles grew to bursting as researchers kept the radical chain growing through ultraviolet light activation. There is, in this, nothing surprising: the forces between the growing vesicle and the surrounding water dictate a critical growth volume before the vesicle ruptures.
The claims should have ended there.
Here is how the work was portrayed in the published article:
The observed net oscillatory vesicle population grows in a manner that reminds one of some elementary modes of sustainable (while there is available “food”!) population growth seen among living systems. The data supports an interpretation in terms of a micron scale self-assembled molecular system capable of embodying and mimicking some aspects of “simple” extant life, including self-assembly from a homogenous but active chemical medium, membrane formation, metabolism, a primitive form of self-replication, and hints of elementary system selection due to a spontaneous light triggered Marangoni instability [provoked by surface tension gradients].4
These claims were then rephrased and presented to the public by the Harvard Gazette:
A Harvard researcher seeking a model for the earliest cells has created a system that self-assembles from a chemical soup into cell-like structures that grow, move in response to light, replicate, and exhibit signs of rudimentary evolutionary selection [emphasis added].5
This degree of hyperbole is excessive.6 Nothing in this experiment had growing cell-like structures with replication, or that exhibited aspects of evolutionary selection.
Teams from the University of California and the University of New South Wales recently conducted lipid bilayer assembly experiments, publishing a summary of their work in 2017.7 They combined nucleotides and lipids in water to form lamellae, with the nucleotides sandwiched between the layers. Nucleotides are trimers of nucleobase-carbohydrate-phosphate, and, in this case, both nucleotides and lipids were purchased in pure homochiral form. Both teams then demonstrated that a condensation polymerization of the nucleotides can take place within the lamella upon dehydration. Polymerization takes place by means of a reaction between pre-loaded phosphate and the purchased stereo-defined alcohol moiety found on a neighboring nucleotide. Similar reactions, they conjectured, may have occurred at the edge of hydrothermal fields, volcanic landmasses providing the necessary heat for reactions.
The chemistry that figures in these experiments is unremarkable. Bear in mind that derivatives were all pre-loaded. To provide the essential concentrations for the reactions, researchers removed the water, thus driving the intermolecular reactions to form oligomers that resembled nucleic acids. The problem with condensation polymerization is obvious: any alcohol can compete for the reactive electrophilic site. In the case under consideration, researchers added no other alcohols. They were scrupulous, but the system was stacked. Condensation polymerization reactions need to be very pure, free of competing nucleophilic and electrophilic components. Witness the Carothers equation, which defines degrees of polymerization based upon monomer purity.8 If there happened to be amino acids or carbohydrates mixed with the nucleotides, they would terminate or interrupt the growth of the oligonucleotides. What is more, the researchers did not confirm the integrity of the structures they claimed to have derived. If carefully analyzed, these structures would likely have shown attacks from unintended hydroxyl sites. Since their sequences are essentially random, short oligonucleotides are not realistic precursors to RNA. An alphabet soup is not a precursor to a poem. The authors go on to suggest that the lamella sandwiching oligonucleotides eventually break off to form lipid bilayer vesicles. These contain the oligonucleotide-within-vesicle constructs, which they call protocells. The conversion of planar lamella into multilamellar vesicles as they hydrate is well established, but shearing forces are generally required to form the requisite lipid bilayer vesicle. For this reason, yields were likely to be low.9 It is hard to imagine finding highly purified homochiral nucleotides trapped in a pure lipid lamella on the prebiotic earth.
But set all that aside. These vesicles bear almost no resemblance to cellular lipid bilayers. Lipid bilayer balls are not cellular lipid bilayers. One would never know this from reading the authors’ account. “Then, in the gel phase,” they write, “protocells pack together in a system called a progenote and exchange sets of polymers, selecting those that enhance survival during many cycles.”10 Chemicals, of course, are indifferent to their survival. No mechanism is described to demonstrate how protocells would bear different sets of polymers or exchange polymers among them. Terms from biology have generally been misappropriated in a way that makes no chemical sense. This is not an isolated or incidental defect. It reappears when the authors write that “[t]he best-adapted protocells spread to other pools or streams, moving by wind and water…”11 Best-adapted? Microbial communities apparently “evolve into a primitive metabolism required by the earliest forms of life.” Molecules do not evolve, and nothing is being metabolized. Condensation polymerization is a simple chemical reaction based upon the addition of nucleophiles to electrophiles with loss of water. Such a reaction is never referred to as a form of metabolism within synthetic chemistry.
Terminology is one thing, non-sequiturs quite another. “After much trial and error,” the authors write, “one protocell assembles the complicated molecular machinery that enables it to divide into daughter cells. This paves the way for the first living microbial community.” How is the molecular machinery made? They do not say. The mechanisms needed for cellular division are complex, requiring cascades of precisely functioning enzymes. There is nothing between what the authors demonstrate and what they claim to have established, and nothing they propose “paves the way for the first living microbial community.”
...
READ MORE HERE: Inference
Você não deve confiar em experimentos que afirmam a existência de universos paralelos
quarta-feira, julho 17, 2019
You Must Not Trust Experiments That Claim The Existence Of Parallel Universes
Ethan Siegel Contributor
Starts With A Bang Contributor Group Science
Ethan Siegel Contributor
Starts With A Bang Contributor Group Science
The Universe is out there, waiting for you to discover it.
A representation of the different parallel
"worlds" that might exist in other pockets of the multiverse, or
anyplace else that theoretical physicists can concoct. Public domain
Is there another Universe out there? The Universe we know and inhabit,
the one that began at the start of the hot Big Bang, might not be the
only one out there. Perhaps one was created at the same time as ours
was, but where time runs backwards instead of forwards. Perhaps there are an infinite number of parallel Universes out there, spawned by an eternally inflating Universe. Or, as has been in the media lately, perhaps there's literally a mirror Universe out there, where the particles we know of are replaced with an exotic version of themselves: mirror matter.
Most scenarios involving parallel Universes like this are untestable, as
we're restricted to living in our own Universe, disconnected from any
others. Yet if one particular idea is right, there might be an experimental signature awaiting our investigations. But even if it yields positive results, you shouldn't trust it. Here's why.
- the BICEP2 collaboration's claimed detection of gravitational waves from inflation,
- the faster-than-light neutrinos claimed from the OPERA experiment,
- or with the diphoton "bump" claimed as evidence for a new particle a few years ago at the LHC.
In all these cases, there was either an error with the way the team did
the analysis or attributed the signal's components, an error in the
experimental setup, or the observed effect was simply a random
statistical fluctuation.
This happens. However, sometimes there are results that really do appear
to be puzzles: the experiments shouldn't turn out the way they did if
the Universe works the way we think it does. These results often turn
out to be omens that we're about to discover new physics, but they also
frequently turn out to be red herrings that lead nowhere. Even worse,
they can turn out to be duds, where they only appear to be interesting
because someone, somewhere, made an error.
...
READ MORE HERE: Forbes
Maquinaria de replicação de DNA capturada em detalhes a nível atômico: mero acaso, fortuita necessidade ou design inteligente?
segunda-feira, julho 15, 2019
DNA translocation mechanism of the MCM complex and implications for replication initiation
Martin Meagher, Leslie B. Epling & Eric J. Enemark
Nature Communications volume 10, Article number: 3117 (2019)
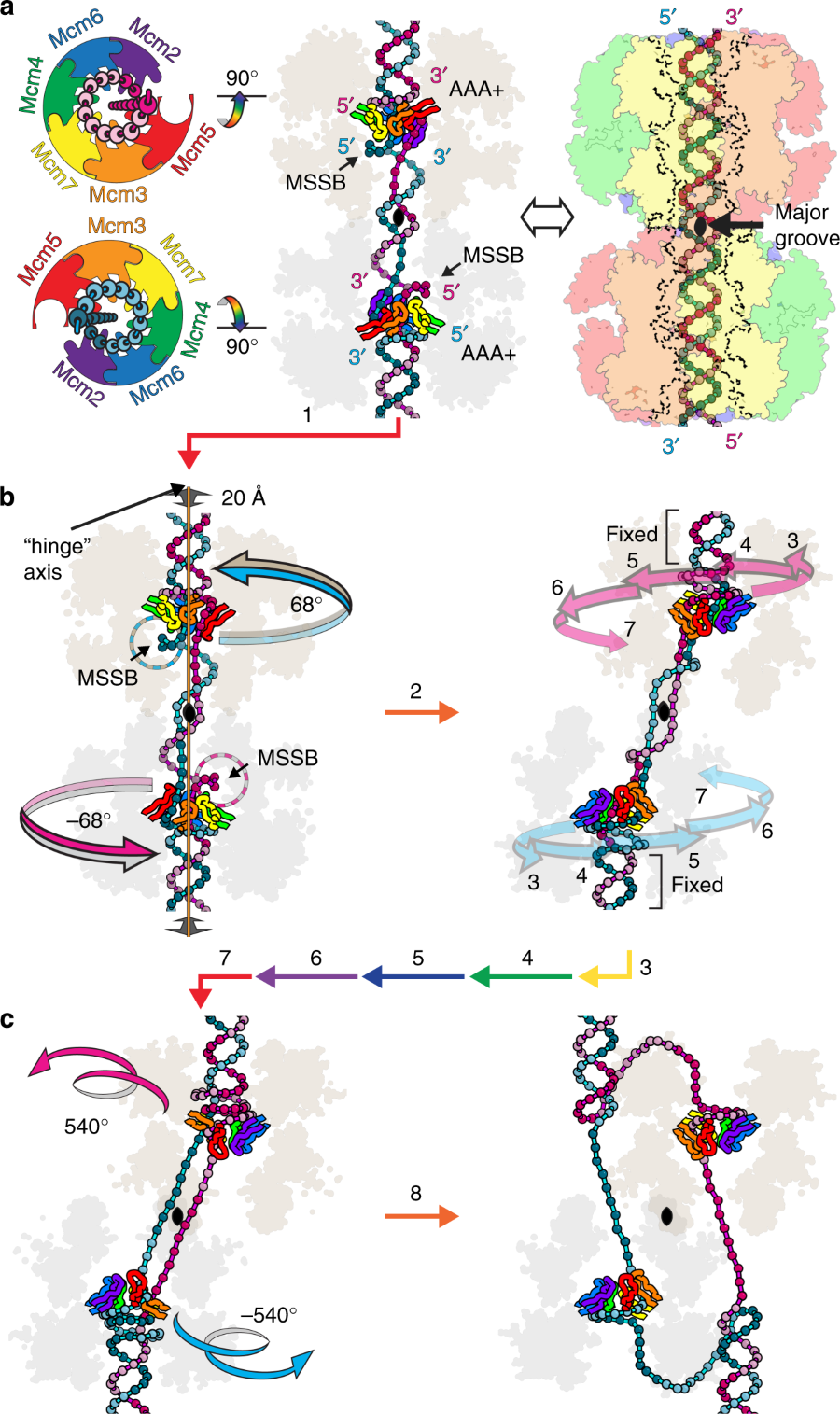
Proposed MCM:DNA aspects of replication initiation.
Abstract
The DNA translocation activity of the minichromosome maintenance (MCM) complex powers DNA strand separation of the replication forks of eukaryotes and archaea. Here we illustrate an atomic level mechanism for this activity with a crystal structure of an archaeal MCM hexamer bound to single-stranded DNA and nucleotide cofactors. Sequence conservation indicates this rotary mechanism is fully possible for all eukaryotes and archaea. The structure definitively demonstrates the ring orients during translocation with the N-terminal domain leading, indicating that the translocation activity could also provide the physical basis of replication initiation where a double-hexamer idly encircling double-stranded DNA transforms to single-hexamers that encircle only one strand. In this mechanism, each strand binds to the N-terminal tier of one hexamer and the AAA+ tier of the other hexamer such that one ring pulls on the other, aligning equivalent interfaces to enable each hexamer to pull its translocation strand outside of the opposing hexamer.
Principais etapas iniciais para a origem da vida ocorrem sob variedade de condições
terça-feira, julho 09, 2019
Nitrogen heterocycles form peptide nucleic acid precursors in complex prebiotic mixtures
Laura E. Rodriguez, Christopher H. House, Karen E. Smith, Melissa R. Roberts & Michael P. Callahan
Scientific Reports volume 9, Article number: 9281 (2019)
Fig. 5: Nitrogen heterocycles form peptide nucleic acid precursors in complex prebiotic mixtures
Abstract
The ability to store information is believed to have been crucial for the origin and evolution of life; however, little is known about the genetic polymers relevant to abiogenesis. Nitrogen heterocycles (N-heterocycles) are plausible components of such polymers as they may have been readily available on early Earth and are the means by which the extant genetic macromolecules RNA and DNA store information. Here, we report the reactivity of numerous N-heterocycles in highly complex mixtures, which were generated using a Miller-Urey spark discharge apparatus with either a reducing or neutral atmosphere, to investigate how N-heterocycles are modified under plausible prebiotic conditions. High throughput mass spectrometry was used to identify N-heterocycle adducts. Additionally, tandem mass spectrometry and nuclear magnetic resonance spectroscopy were used to elucidate reaction pathways for select reactions. Remarkably, we found that the majority of N-heterocycles, including the canonical nucleobases, gain short carbonyl side chains in our complex mixtures via a Strecker-like synthesis or Michael addition. These types of N-heterocycle adducts are subunits of the proposed RNA precursor, peptide nucleic acids (PNAs). The ease with which these carbonylated heterocycles form under both reducing and neutral atmospheres is suggestive that PNAs could be prebiotically feasible on early Earth.
FREE PDF GRATIS: Scientific Reports Sup. Info.
Mutações: O engano da evolução X-Men
segunda-feira, julho 08, 2019
#ScienceUprising #RevoltaDaCiencia
Darwin, mais complexidade: microscopia de super resolução ilumina as associações entre os cromossomos
quinta-feira, julho 04, 2019
Superresolution microscopy reveals linkages between ribosomal DNA on heterologous chromosomes
DOI: 10.1083/jcb.201810166 | Published July 3, 2019
FREE PDF GRATIS: Journal of Cell Biology
Tamara A. Potapova, Jay R. Unruh, Zulin Yu, Giulia Rancati, Hua Li, Martha R. Stampfer, Jennifer L. Gerton
DOI: 10.1083/jcb.201810166 | Published July 3, 2019
Abstract
The spatial organization of the genome is enigmatic. Direct evidence of physical contacts between chromosomes and their visualization at nanoscale resolution has been limited. We used superresolution microscopy to demonstrate that ribosomal DNA (rDNA) can form linkages between chromosomes. We observed rDNA linkages in many different human cell types and demonstrated their resolution in anaphase. rDNA linkages are coated by the transcription factor UBF and their formation depends on UBF, indicating that they regularly occur between transcriptionally active loci. Overexpression of c-Myc increases rDNA transcription and the frequency of rDNA linkages, further suggesting that their formation depends on active transcription. Linkages persist in the absence of cohesion, but inhibition of topoisomerase II prevents their resolution in anaphase. We propose that linkages are topological intertwines occurring between transcriptionally active rDNA loci spatially colocated in the same nucleolar compartment. Our findings suggest that active DNA loci engage in physical interchromosomal connections that are an integral and pervasive feature of genome organization.
The spatial organization of the genome is enigmatic. Direct evidence of physical contacts between chromosomes and their visualization at nanoscale resolution has been limited. We used superresolution microscopy to demonstrate that ribosomal DNA (rDNA) can form linkages between chromosomes. We observed rDNA linkages in many different human cell types and demonstrated their resolution in anaphase. rDNA linkages are coated by the transcription factor UBF and their formation depends on UBF, indicating that they regularly occur between transcriptionally active loci. Overexpression of c-Myc increases rDNA transcription and the frequency of rDNA linkages, further suggesting that their formation depends on active transcription. Linkages persist in the absence of cohesion, but inhibition of topoisomerase II prevents their resolution in anaphase. We propose that linkages are topological intertwines occurring between transcriptionally active rDNA loci spatially colocated in the same nucleolar compartment. Our findings suggest that active DNA loci engage in physical interchromosomal connections that are an integral and pervasive feature of genome organization.
Origem da vida: requer inteligência
segunda-feira, julho 01, 2019
#ScienceUprising #RevoltaDaCiencia
Assinar:
Comentários (Atom)






