Translation affects mRNA stability in a codon-dependent manner in human cells
Qiushuang Wu, Santiago Gerardo Medina, Gopal Kushawah, Michelle Lynn DeVore, Luciana A Castellano, Jacqelyn M Hand, Matthew Wright, Ariel Alejandro Bazzini
Stowers Institute for Medical Research, United States
RESEARCH ARTICLE Apr 23, 2019
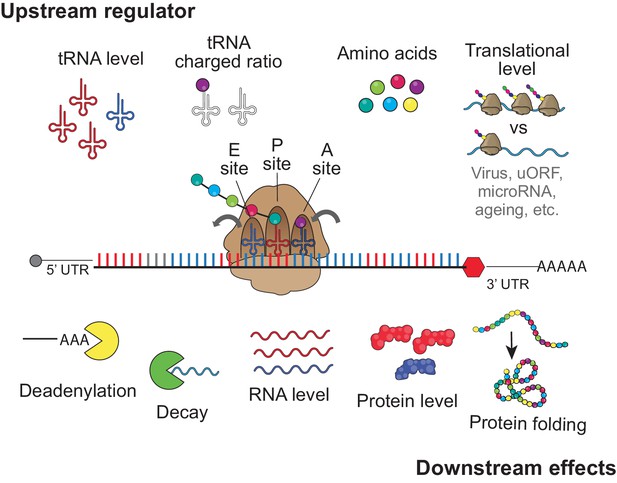
Upstream regulator and downstream effect for codon optimality tRNA level, tRNA charged ratio, amino acid, and translational level might contribute to regulates the regulatory identity and/or strength of each codon to affect gene expression, by influencing the speed of translation elongation.
Abstract
mRNA translation decodes nucleotide into amino acid sequences. However, translation has also been shown to affect mRNA stability depending on codon composition in model organisms, although universality of this mechanism remains unclear. Here, using three independent approaches to measure exogenous and endogenous mRNA decay, we define which codons are associated with stable or unstable mRNAs in human cells. We demonstrate that the regulatory information affecting mRNA stability is encoded in codons and not in nucleotides. Stabilizing codons tend to be associated with higher tRNA levels and higher charged/total tRNA ratios. While mRNAs enriched in destabilizing codons tend to possess shorter poly(A)-tails, the poly(A)-tail is not required for the codon-mediated mRNA stability. This mechanism depends on translation; however, the number of ribosome loads into a mRNA modulates the codon-mediated effects on gene expression. This work provides definitive evidence that translation strongly affects mRNA stability in a codon-dependent manner in human cells.
https://doi.org/10.7554/eLife.45396.001
eLife digest
Proteins are made by joining together building blocks called amino acids into strings. The proteins are ‘translated’ from genetic sequences called mRNA molecules. These sequences can be thought of as series of ‘letters’, which are read in groups of three known as codons. Molecules called tRNAs recognize the codons and add the matching amino acids to the end of the protein. Each tRNA can recognize one or several codons, and the levels of different tRNAs inside the cell vary.
There are 61 codons that code for amino acids, but only 20 amino acids. This means that some codons produce the same amino acid. Despite this, there is evidence to suggest that not all of the codons that produce the same amino acid are exactly equivalent. In bacteria, yeast and zebrafish, some codons seem to make the mRNA molecule more stable, and others make it less stable. This might help the cell to control how many proteins it makes. It was not clear whether the same is true for humans.
To find out, Wu et al. used three separate methods to examine mRNA stability in four types of human cell. Overall, the results revealed that some codons help to stabilize the mRNA, while others make the mRNA molecule break down faster. The effect seems to depend on the supply of tRNAs that have a charged amino acid; mRNA molecules were more likely to self-destruct in cells that contained codons with low levels of the tRNA molecules.
Wu et al. also found that conditions in the cell can alter how strongly the codons affect mRNA stability. For example, a cell that has been infected by a virus reduces translation. Under these conditions, the identity of the codons in the mRNA has less effect on the stability of the mRNA molecule.
Changes to protein production happen in many diseases. Understanding what controls these changes could help to reveal more about our fundamental biology, and what happens when it goes wrong.
https://doi.org/10.7554/eLife.45396.002
FREE PDF GRATIS: eLIFE


