REVIEW ARTICLE
First Detailed Anatomical Study of Bonobos Reveals Intra-Specific Variations and Exposes Just-So Stories of Human Evolution, Bipedalism, and Tool Use
Rui Diogo*
Department of Anatomy, Howard University, Washington, DC, United States
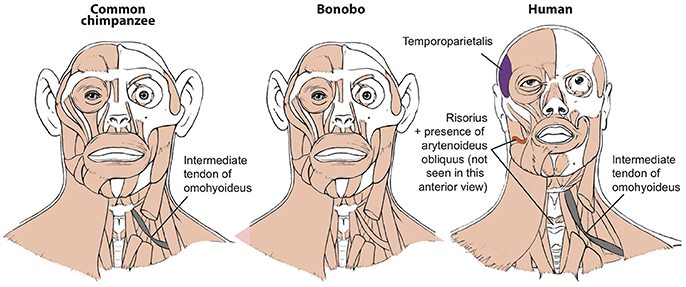
Figure 1. Differences between head muscles of common chimps, bonobos, and humans, based and modified from Diogo et al. (2017b).
Just-so stories are prominent in human evolution literature because of our tendency to create simple progressionist narratives about our “special” place in nature, despite the fact that these stories are almost exclusively based on hard tissue data. How can we be so certain about the evolution of human facial communication, bipedalism, tool use, or speech without detailed knowledge of the internal anatomy of for instance, one of the two extant species more closely related to us, the bonobos? Here I show how many of these stories now become obsolete, after such a comprehensive knowledge on the anatomy of bonobos and other primates is finally put together. Each and every muscle that has been long accepted to be “uniquely human” and to provide “crucial singular functional adaptations” for our bipedalism, tool use and/or vocal/facial communication, is actually present as an intra-specific variant or even as normal phenotype in bonobos and/or other apes.
Just-so stories (Smith, 2016) are frequent in the literature about human evolution because of our tendency to build simple progressionist narratives about our “special” evolutionary history and place in nature (Gould, 1993, 2002). This is particularly striking because these stories are in reality almost exclusively based on hard tissue data. In fact, descriptions of the soft tissues of apes have been relatively scarce and mainly referred to just a few muscles of the head or limbs of a single taxon, in most cases (e.g., Tyson, 1699; Bischoff, 1880; Raven, 1950; Swindler and Wood, 1973; Diogo and Wood, 2011, 2012; Persaud and Loukas, 2014). For instance, the only study that specifically focused on the musculature of bonobos (Pan paniscus) was that of Miller (1952), which was based on dissections of a single adult and did not provide information for numerous head and limb muscles (Diogo and Wood, 2011, 2012). Strikingly, despite this scarcity of information, biologists and anthropologists have displayed a remarkable confidence in their stories about the origin and evolution of human soft tissues, including their phylogenetic distribution and “singular functional adaptations.”
To illustrate this fact, in this short paper I will refer here briefly to seven muscles that have long been generally seen as “unique human features” and linked with specific adaptations for our bipedalism, tool use, and vocal or facial communication. Firstly, the facial expression muscle risorius (Figure 1) has been generally accepted as a unique feature crucial for the evolution of our “gracile” smile and “specially sophisticated” facial communication abilities (Huber, 1931). In a very influential paper, Susman et al. argued—although (fortunately) not as confidently as the assertions done by some of the other authors cited here—that the hand muscle adductor pollicis accessorius (Figure 2; “Henle” or “interosseous volaris primus” muscle: Bello-Hellegouarch et al., 2013) is a unique feature likely related to our increased ability to manufacture and/or use tools (Susman et al., 1999). Similarly, the foot muscle adductor hallucis accessorius—which topologically corresponds to the adductor pollicis accessorius of the hand—is also often considered to be uniquely found in our bipedal species, being at least consistently present at early stages of our ontogenetic development (Cihak, 1972). The foot muscle fibularis tertius (Figure 3) is, according to Lewis' (1989) highly influential monograph on the evolution of our limbs, a unique feature most likely associated with our bipedal evolution (Lewis, 1989). The flexor pollicis longus and extensor pollicis brevis (Figure 2) are forearm muscles that insert onto the thumb and that are generally considered to be unique adaptations for human tool manufacture and use (Lewis, 1989). For instance, it has been experimentally shown that the recruitment to these two muscles allows human subjects to maintain the metacarpophalangeal joint in extension while flexing the distal phalanx of the thumb, i.e., two primary movements usually done when we grab/manipulate objects (Marzke et al., 1998; Williams et al., 2012). Lastly, the laryngeal muscle arytenoideus obliquus has long been considered to be a unique feature of humans—which also have an arytenoideus transversus, in contrast to the single arytenoideus muscle said to occur in all other primates—associated to our enhanced vocal communication (reviewed in Diogo and Wood, 2012).